
Modestobacter caceresii
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Modestobacter
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
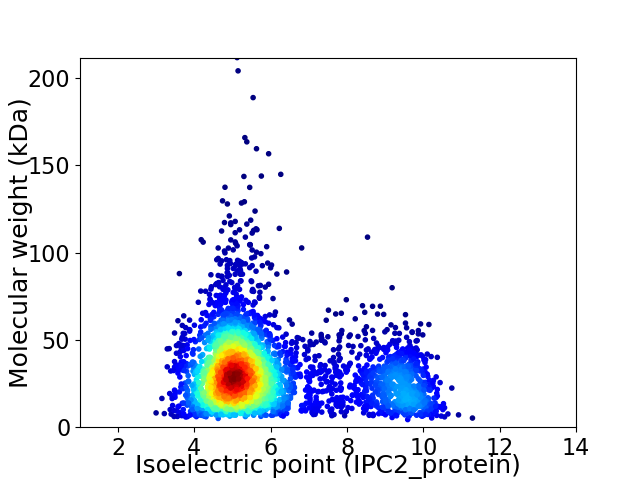
Virtual 2D-PAGE plot for 3847 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A098Y9H3|A0A098Y9H3_9ACTN Methylase OS=Modestobacter caceresii OX=1522368 GN=IN07_08945 PE=4 SV=1
MM1 pKa = 7.35NSVSRR6 pKa = 11.84SGRR9 pKa = 11.84RR10 pKa = 11.84ALVATAGITATALALSACGGDD31 pKa = 4.16DD32 pKa = 3.7GDD34 pKa = 4.7GGGSGGGDD42 pKa = 3.33SLIIGTTDD50 pKa = 3.59KK51 pKa = 10.43ITTIDD56 pKa = 3.49PAGSYY61 pKa = 11.17DD62 pKa = 3.54NGSFAVMNQVYY73 pKa = 8.97PFLMNTPLGSPDD85 pKa = 3.76VEE87 pKa = 3.9PDD89 pKa = 3.01IAEE92 pKa = 4.25SAEE95 pKa = 4.09FTSPTEE101 pKa = 3.89YY102 pKa = 10.56TVTLKK107 pKa = 11.03DD108 pKa = 3.31GLTFANGNEE117 pKa = 4.04LTASDD122 pKa = 4.03VKK124 pKa = 10.89FSFDD128 pKa = 3.5RR129 pKa = 11.84MVAINDD135 pKa = 3.42EE136 pKa = 4.45SGPASLLYY144 pKa = 10.33NLEE147 pKa = 4.04STEE150 pKa = 4.16VVDD153 pKa = 3.97DD154 pKa = 3.79TTVVFNLKK162 pKa = 10.62SPDD165 pKa = 3.66DD166 pKa = 3.65QVFPQILSSPAGPIVDD182 pKa = 3.9EE183 pKa = 5.08DD184 pKa = 4.14VFAADD189 pKa = 5.75ALTTDD194 pKa = 3.78QEE196 pKa = 4.56IVDD199 pKa = 4.23GNAFAGPYY207 pKa = 10.29AITNYY212 pKa = 10.11DD213 pKa = 3.34QNNLVSYY220 pKa = 9.39EE221 pKa = 4.0AFEE224 pKa = 5.05GYY226 pKa = 9.63QGLLGAPEE234 pKa = 4.23TEE236 pKa = 4.55TVNVRR241 pKa = 11.84YY242 pKa = 9.58YY243 pKa = 11.63ADD245 pKa = 3.67ASNLKK250 pKa = 10.44LDD252 pKa = 3.86VQEE255 pKa = 4.86GNVDD259 pKa = 3.42VAFRR263 pKa = 11.84SLTATDD269 pKa = 3.86VEE271 pKa = 4.73DD272 pKa = 4.47LRR274 pKa = 11.84GDD276 pKa = 3.56DD277 pKa = 3.67SVNVVDD283 pKa = 5.57GPGGEE288 pKa = 3.65IRR290 pKa = 11.84YY291 pKa = 8.28ITFNFNTQPYY301 pKa = 9.66GATTPEE307 pKa = 4.22ADD309 pKa = 3.52PAKK312 pKa = 10.57ALAVRR317 pKa = 11.84QAAAHH322 pKa = 6.45LIDD325 pKa = 4.53RR326 pKa = 11.84EE327 pKa = 4.06EE328 pKa = 4.85LADD331 pKa = 3.59QVYY334 pKa = 10.44KK335 pKa = 10.17GTYY338 pKa = 7.53TPLYY342 pKa = 9.34SYY344 pKa = 10.97VPEE347 pKa = 4.54GLTGAVEE354 pKa = 4.13PLLEE358 pKa = 4.61KK359 pKa = 11.22YY360 pKa = 10.72GDD362 pKa = 3.78GSGAPDD368 pKa = 3.58AEE370 pKa = 4.26AAAQVLQAAGVQTPVEE386 pKa = 4.26LNLQYY391 pKa = 11.47SNDD394 pKa = 3.55HH395 pKa = 6.32YY396 pKa = 11.31GPSSGDD402 pKa = 3.07EE403 pKa = 3.82YY404 pKa = 11.92ALIKK408 pKa = 10.69DD409 pKa = 3.71QLEE412 pKa = 4.2SSGLFTVNLQTTEE425 pKa = 3.68WVQYY429 pKa = 10.88SEE431 pKa = 5.65DD432 pKa = 3.69RR433 pKa = 11.84TADD436 pKa = 3.37VYY438 pKa = 10.68PAYY441 pKa = 10.26QLGWFPDD448 pKa = 3.67YY449 pKa = 11.48SDD451 pKa = 4.11ADD453 pKa = 3.57NYY455 pKa = 8.96LTPFFLTEE463 pKa = 3.86NFLGNHH469 pKa = 5.69YY470 pKa = 10.65SDD472 pKa = 5.11PEE474 pKa = 4.26VNDD477 pKa = 5.67LILQQATTTDD487 pKa = 3.61PAEE490 pKa = 4.2RR491 pKa = 11.84EE492 pKa = 3.98ALIEE496 pKa = 4.02QIQGQVADD504 pKa = 4.97DD505 pKa = 4.92LSTLPYY511 pKa = 10.41LQGAQVAVTGTDD523 pKa = 3.25VQGAEE528 pKa = 4.25DD529 pKa = 3.78TLDD532 pKa = 3.46PSFKK536 pKa = 10.15FRR538 pKa = 11.84YY539 pKa = 9.04GALSKK544 pKa = 11.17GG545 pKa = 3.56
MM1 pKa = 7.35NSVSRR6 pKa = 11.84SGRR9 pKa = 11.84RR10 pKa = 11.84ALVATAGITATALALSACGGDD31 pKa = 4.16DD32 pKa = 3.7GDD34 pKa = 4.7GGGSGGGDD42 pKa = 3.33SLIIGTTDD50 pKa = 3.59KK51 pKa = 10.43ITTIDD56 pKa = 3.49PAGSYY61 pKa = 11.17DD62 pKa = 3.54NGSFAVMNQVYY73 pKa = 8.97PFLMNTPLGSPDD85 pKa = 3.76VEE87 pKa = 3.9PDD89 pKa = 3.01IAEE92 pKa = 4.25SAEE95 pKa = 4.09FTSPTEE101 pKa = 3.89YY102 pKa = 10.56TVTLKK107 pKa = 11.03DD108 pKa = 3.31GLTFANGNEE117 pKa = 4.04LTASDD122 pKa = 4.03VKK124 pKa = 10.89FSFDD128 pKa = 3.5RR129 pKa = 11.84MVAINDD135 pKa = 3.42EE136 pKa = 4.45SGPASLLYY144 pKa = 10.33NLEE147 pKa = 4.04STEE150 pKa = 4.16VVDD153 pKa = 3.97DD154 pKa = 3.79TTVVFNLKK162 pKa = 10.62SPDD165 pKa = 3.66DD166 pKa = 3.65QVFPQILSSPAGPIVDD182 pKa = 3.9EE183 pKa = 5.08DD184 pKa = 4.14VFAADD189 pKa = 5.75ALTTDD194 pKa = 3.78QEE196 pKa = 4.56IVDD199 pKa = 4.23GNAFAGPYY207 pKa = 10.29AITNYY212 pKa = 10.11DD213 pKa = 3.34QNNLVSYY220 pKa = 9.39EE221 pKa = 4.0AFEE224 pKa = 5.05GYY226 pKa = 9.63QGLLGAPEE234 pKa = 4.23TEE236 pKa = 4.55TVNVRR241 pKa = 11.84YY242 pKa = 9.58YY243 pKa = 11.63ADD245 pKa = 3.67ASNLKK250 pKa = 10.44LDD252 pKa = 3.86VQEE255 pKa = 4.86GNVDD259 pKa = 3.42VAFRR263 pKa = 11.84SLTATDD269 pKa = 3.86VEE271 pKa = 4.73DD272 pKa = 4.47LRR274 pKa = 11.84GDD276 pKa = 3.56DD277 pKa = 3.67SVNVVDD283 pKa = 5.57GPGGEE288 pKa = 3.65IRR290 pKa = 11.84YY291 pKa = 8.28ITFNFNTQPYY301 pKa = 9.66GATTPEE307 pKa = 4.22ADD309 pKa = 3.52PAKK312 pKa = 10.57ALAVRR317 pKa = 11.84QAAAHH322 pKa = 6.45LIDD325 pKa = 4.53RR326 pKa = 11.84EE327 pKa = 4.06EE328 pKa = 4.85LADD331 pKa = 3.59QVYY334 pKa = 10.44KK335 pKa = 10.17GTYY338 pKa = 7.53TPLYY342 pKa = 9.34SYY344 pKa = 10.97VPEE347 pKa = 4.54GLTGAVEE354 pKa = 4.13PLLEE358 pKa = 4.61KK359 pKa = 11.22YY360 pKa = 10.72GDD362 pKa = 3.78GSGAPDD368 pKa = 3.58AEE370 pKa = 4.26AAAQVLQAAGVQTPVEE386 pKa = 4.26LNLQYY391 pKa = 11.47SNDD394 pKa = 3.55HH395 pKa = 6.32YY396 pKa = 11.31GPSSGDD402 pKa = 3.07EE403 pKa = 3.82YY404 pKa = 11.92ALIKK408 pKa = 10.69DD409 pKa = 3.71QLEE412 pKa = 4.2SSGLFTVNLQTTEE425 pKa = 3.68WVQYY429 pKa = 10.88SEE431 pKa = 5.65DD432 pKa = 3.69RR433 pKa = 11.84TADD436 pKa = 3.37VYY438 pKa = 10.68PAYY441 pKa = 10.26QLGWFPDD448 pKa = 3.67YY449 pKa = 11.48SDD451 pKa = 4.11ADD453 pKa = 3.57NYY455 pKa = 8.96LTPFFLTEE463 pKa = 3.86NFLGNHH469 pKa = 5.69YY470 pKa = 10.65SDD472 pKa = 5.11PEE474 pKa = 4.26VNDD477 pKa = 5.67LILQQATTTDD487 pKa = 3.61PAEE490 pKa = 4.2RR491 pKa = 11.84EE492 pKa = 3.98ALIEE496 pKa = 4.02QIQGQVADD504 pKa = 4.97DD505 pKa = 4.92LSTLPYY511 pKa = 10.41LQGAQVAVTGTDD523 pKa = 3.25VQGAEE528 pKa = 4.25DD529 pKa = 3.78TLDD532 pKa = 3.46PSFKK536 pKa = 10.15FRR538 pKa = 11.84YY539 pKa = 9.04GALSKK544 pKa = 11.17GG545 pKa = 3.56
Molecular weight: 58.25 kDa
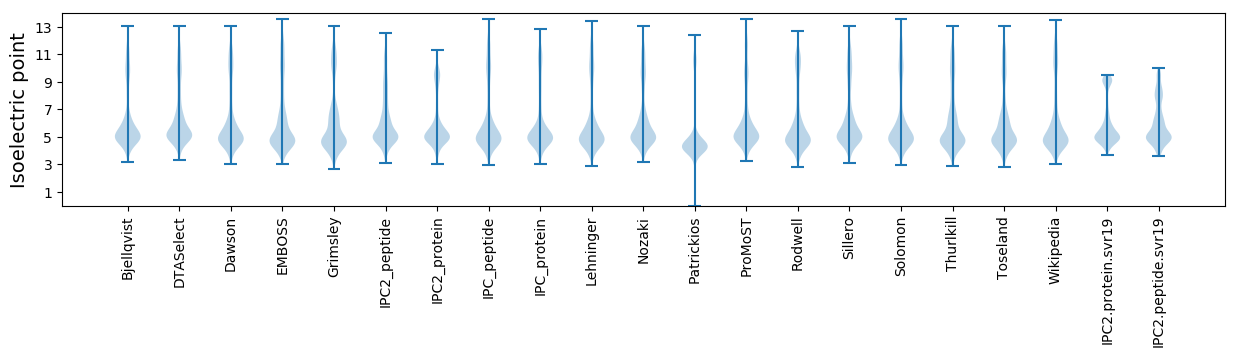
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A098YDQ2|A0A098YDQ2_9ACTN Bifunctional purine biosynthesis protein PurH OS=Modestobacter caceresii OX=1522368 GN=purH PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPNTRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.83THH17 pKa = 5.18GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.23GRR40 pKa = 11.84NKK42 pKa = 10.44LSAA45 pKa = 3.84
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPNTRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.83THH17 pKa = 5.18GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.23GRR40 pKa = 11.84NKK42 pKa = 10.44LSAA45 pKa = 3.84
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1186502 |
37 |
2005 |
308.4 |
32.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.24 ± 0.066 | 0.666 ± 0.01 |
6.286 ± 0.034 | 5.636 ± 0.031 |
2.563 ± 0.023 | 9.522 ± 0.031 |
2.037 ± 0.021 | 2.83 ± 0.034 |
1.424 ± 0.026 | 10.797 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.648 ± 0.015 | 1.471 ± 0.019 |
6.03 ± 0.033 | 2.904 ± 0.02 |
7.839 ± 0.045 | 4.891 ± 0.024 |
6.128 ± 0.027 | 9.86 ± 0.038 |
1.484 ± 0.018 | 1.746 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
