
Sulfolobus monocaudavirus SMV2
Taxonomy: Viruses; Bicaudaviridae; unclassified Bicaudaviridae
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
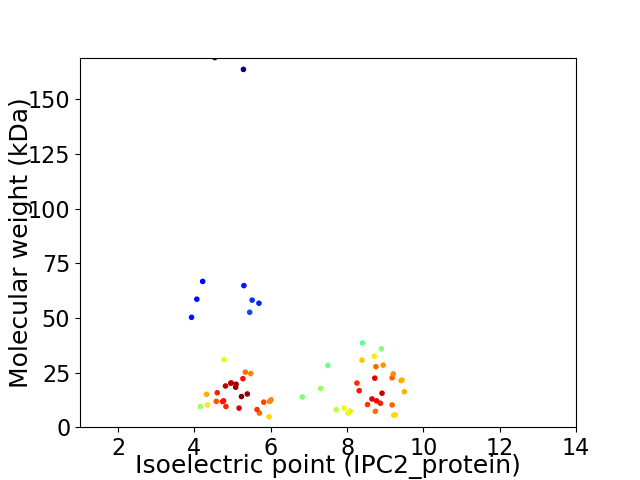
Virtual 2D-PAGE plot for 65 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0N7A9S6|A0A0N7A9S6_9VIRU MoxR-like ATPase OS=Sulfolobus monocaudavirus SMV2 OX=1580591 PE=4 SV=1
MM1 pKa = 7.21SQSQKK6 pKa = 10.61PNLYY10 pKa = 10.38LLSLYY15 pKa = 9.24PSPVGVTVTGDD26 pKa = 3.33TTDD29 pKa = 3.29QTVSFVISPYY39 pKa = 9.75HH40 pKa = 5.69YY41 pKa = 11.04VDD43 pKa = 3.15ISYY46 pKa = 10.73LITSYY51 pKa = 11.63NDD53 pKa = 3.33GVQVYY58 pKa = 9.81APDD61 pKa = 3.86ANIPQSGISGFPPGVPTVTLIIEE84 pKa = 4.34KK85 pKa = 10.73GNLNFEE91 pKa = 4.39FATPEE96 pKa = 4.97DD97 pKa = 3.5ILKK100 pKa = 10.65SYY102 pKa = 10.45LYY104 pKa = 11.24NNILQNMNNDD114 pKa = 3.12LTEE117 pKa = 4.54IYY119 pKa = 10.22QDD121 pKa = 4.81LITLISEE128 pKa = 4.45YY129 pKa = 11.18ASGQSLSQSYY139 pKa = 10.34LSEE142 pKa = 4.14VSSLLQQINNMLQNAEE158 pKa = 4.08QFEE161 pKa = 4.49NTANLSFEE169 pKa = 4.41GVSALQQVYY178 pKa = 11.1NNLNEE183 pKa = 4.42IYY185 pKa = 10.67NSLTNQTLTLGEE197 pKa = 4.55LEE199 pKa = 4.35SLQLPSYY206 pKa = 7.54STPEE210 pKa = 3.76ASSIASNYY218 pKa = 7.33TQFLNNAVSIMQNASSQSGSSSSSSQGSNSSSSTPQSSSSSNASSQSGSSSSPSPLSSLTTNSQSGTSSSSYY290 pKa = 8.96NTQTSTSSTTTITTSTSTSSTTSQTSIISPPTLSQSQVLAQIQAGDD336 pKa = 3.69VSSLLSEE343 pKa = 4.34LSEE346 pKa = 4.45IPSNMIPVVNTVVQLYY362 pKa = 9.36QLYY365 pKa = 9.52GFPDD369 pKa = 4.34GTPLSTVVEE378 pKa = 4.23RR379 pKa = 11.84LLSDD383 pKa = 3.12IRR385 pKa = 11.84EE386 pKa = 3.99AYY388 pKa = 9.85YY389 pKa = 10.86NIVNHH394 pKa = 5.84KK395 pKa = 9.46QEE397 pKa = 4.33EE398 pKa = 4.53VNLQQLSQLLQLYY411 pKa = 7.42NTLVSNGLAPQSQSASKK428 pKa = 10.57LSEE431 pKa = 3.87LTQTSEE437 pKa = 5.04IIPALPPVLGQPYY450 pKa = 9.16PGRR453 pKa = 11.84VNSNSNSNSVNYY465 pKa = 10.84SNFII469 pKa = 3.77
MM1 pKa = 7.21SQSQKK6 pKa = 10.61PNLYY10 pKa = 10.38LLSLYY15 pKa = 9.24PSPVGVTVTGDD26 pKa = 3.33TTDD29 pKa = 3.29QTVSFVISPYY39 pKa = 9.75HH40 pKa = 5.69YY41 pKa = 11.04VDD43 pKa = 3.15ISYY46 pKa = 10.73LITSYY51 pKa = 11.63NDD53 pKa = 3.33GVQVYY58 pKa = 9.81APDD61 pKa = 3.86ANIPQSGISGFPPGVPTVTLIIEE84 pKa = 4.34KK85 pKa = 10.73GNLNFEE91 pKa = 4.39FATPEE96 pKa = 4.97DD97 pKa = 3.5ILKK100 pKa = 10.65SYY102 pKa = 10.45LYY104 pKa = 11.24NNILQNMNNDD114 pKa = 3.12LTEE117 pKa = 4.54IYY119 pKa = 10.22QDD121 pKa = 4.81LITLISEE128 pKa = 4.45YY129 pKa = 11.18ASGQSLSQSYY139 pKa = 10.34LSEE142 pKa = 4.14VSSLLQQINNMLQNAEE158 pKa = 4.08QFEE161 pKa = 4.49NTANLSFEE169 pKa = 4.41GVSALQQVYY178 pKa = 11.1NNLNEE183 pKa = 4.42IYY185 pKa = 10.67NSLTNQTLTLGEE197 pKa = 4.55LEE199 pKa = 4.35SLQLPSYY206 pKa = 7.54STPEE210 pKa = 3.76ASSIASNYY218 pKa = 7.33TQFLNNAVSIMQNASSQSGSSSSSSQGSNSSSSTPQSSSSSNASSQSGSSSSPSPLSSLTTNSQSGTSSSSYY290 pKa = 8.96NTQTSTSSTTTITTSTSTSSTTSQTSIISPPTLSQSQVLAQIQAGDD336 pKa = 3.69VSSLLSEE343 pKa = 4.34LSEE346 pKa = 4.45IPSNMIPVVNTVVQLYY362 pKa = 9.36QLYY365 pKa = 9.52GFPDD369 pKa = 4.34GTPLSTVVEE378 pKa = 4.23RR379 pKa = 11.84LLSDD383 pKa = 3.12IRR385 pKa = 11.84EE386 pKa = 3.99AYY388 pKa = 9.85YY389 pKa = 10.86NIVNHH394 pKa = 5.84KK395 pKa = 9.46QEE397 pKa = 4.33EE398 pKa = 4.53VNLQQLSQLLQLYY411 pKa = 7.42NTLVSNGLAPQSQSASKK428 pKa = 10.57LSEE431 pKa = 3.87LTQTSEE437 pKa = 5.04IIPALPPVLGQPYY450 pKa = 9.16PGRR453 pKa = 11.84VNSNSNSNSVNYY465 pKa = 10.84SNFII469 pKa = 3.77
Molecular weight: 50.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0N7AA29|A0A0N7AA29_9VIRU Uncharacterized protein OS=Sulfolobus monocaudavirus SMV2 OX=1580591 PE=4 SV=1
MM1 pKa = 8.29VEE3 pKa = 3.94VKK5 pKa = 10.57LRR7 pKa = 11.84WRR9 pKa = 11.84VKK11 pKa = 9.94INRR14 pKa = 11.84KK15 pKa = 8.62NGKK18 pKa = 9.4EE19 pKa = 3.65YY20 pKa = 9.03MLRR23 pKa = 11.84YY24 pKa = 9.84INVTSRR30 pKa = 11.84SAVFLINYY38 pKa = 8.13VPYY41 pKa = 10.69FNIKK45 pKa = 10.32NNIIEE50 pKa = 4.43FKK52 pKa = 10.14PNRR55 pKa = 11.84RR56 pKa = 11.84YY57 pKa = 10.0AKK59 pKa = 9.15KK60 pKa = 10.12GLRR63 pKa = 11.84IRR65 pKa = 11.84LRR67 pKa = 11.84WLIKK71 pKa = 10.0RR72 pKa = 11.84EE73 pKa = 3.97KK74 pKa = 10.65KK75 pKa = 9.57NGKK78 pKa = 9.14VFTQYY83 pKa = 10.79YY84 pKa = 9.57ISIPSEE90 pKa = 3.61LAKK93 pKa = 10.46PVVDD97 pKa = 4.91LDD99 pKa = 4.14PYY101 pKa = 10.93LVPEE105 pKa = 3.94NMQIVFRR112 pKa = 11.84PKK114 pKa = 10.2EE115 pKa = 3.92GDD117 pKa = 3.54GMGLNNQLFVDD128 pKa = 4.98DD129 pKa = 4.64EE130 pKa = 5.33GEE132 pKa = 4.31TQSQQ136 pKa = 3.19
MM1 pKa = 8.29VEE3 pKa = 3.94VKK5 pKa = 10.57LRR7 pKa = 11.84WRR9 pKa = 11.84VKK11 pKa = 9.94INRR14 pKa = 11.84KK15 pKa = 8.62NGKK18 pKa = 9.4EE19 pKa = 3.65YY20 pKa = 9.03MLRR23 pKa = 11.84YY24 pKa = 9.84INVTSRR30 pKa = 11.84SAVFLINYY38 pKa = 8.13VPYY41 pKa = 10.69FNIKK45 pKa = 10.32NNIIEE50 pKa = 4.43FKK52 pKa = 10.14PNRR55 pKa = 11.84RR56 pKa = 11.84YY57 pKa = 10.0AKK59 pKa = 9.15KK60 pKa = 10.12GLRR63 pKa = 11.84IRR65 pKa = 11.84LRR67 pKa = 11.84WLIKK71 pKa = 10.0RR72 pKa = 11.84EE73 pKa = 3.97KK74 pKa = 10.65KK75 pKa = 9.57NGKK78 pKa = 9.14VFTQYY83 pKa = 10.79YY84 pKa = 9.57ISIPSEE90 pKa = 3.61LAKK93 pKa = 10.46PVVDD97 pKa = 4.91LDD99 pKa = 4.14PYY101 pKa = 10.93LVPEE105 pKa = 3.94NMQIVFRR112 pKa = 11.84PKK114 pKa = 10.2EE115 pKa = 3.92GDD117 pKa = 3.54GMGLNNQLFVDD128 pKa = 4.98DD129 pKa = 4.64EE130 pKa = 5.33GEE132 pKa = 4.31TQSQQ136 pKa = 3.19
Molecular weight: 16.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14878 |
45 |
1563 |
228.9 |
25.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.807 ± 0.382 | 0.376 ± 0.085 |
4.315 ± 0.313 | 6.701 ± 0.672 |
4.463 ± 0.319 | 5.175 ± 0.318 |
0.921 ± 0.09 | 7.783 ± 0.309 |
6.466 ± 0.631 | 10.075 ± 0.333 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.124 ± 0.191 | 6.083 ± 0.399 |
4.241 ± 0.329 | 4.651 ± 0.657 |
3.32 ± 0.359 | 7.494 ± 0.582 |
5.767 ± 0.46 | 7.151 ± 0.395 |
0.86 ± 0.141 | 5.29 ± 0.363 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
