
Thermobifida halotolerans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Nocardiopsaceae; Thermobifida
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
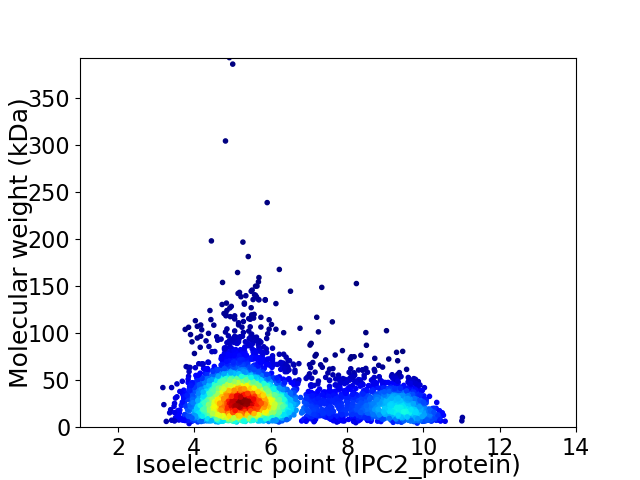
Virtual 2D-PAGE plot for 4067 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A399G272|A0A399G272_9ACTN DNA helicase OS=Thermobifida halotolerans OX=483545 GN=NI17_15575 PE=3 SV=1
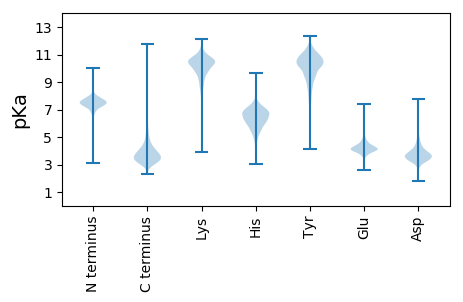
MM1 pKa = 7.26SKK3 pKa = 10.02VRR5 pKa = 11.84ATNRR9 pKa = 11.84RR10 pKa = 11.84SWLRR14 pKa = 11.84RR15 pKa = 11.84GFATASSLALGASMVALAAPANAAGCSVDD44 pKa = 3.52YY45 pKa = 10.63QVNSWGSGFTASVTITNLGSALSNWTLEE73 pKa = 3.46WDD75 pKa = 4.02FPGNQQVTNLWNGSYY90 pKa = 9.81SQSGQHH96 pKa = 5.44VSVDD100 pKa = 3.3NAPYY104 pKa = 9.93NGSVGEE110 pKa = 4.25GGTVNFGFNGSYY122 pKa = 10.11SGSNDD127 pKa = 2.6IPTSFSLNGVTCDD140 pKa = 3.94GSVDD144 pKa = 4.01PDD146 pKa = 4.08PPTDD150 pKa = 3.7PTDD153 pKa = 3.95PPTDD157 pKa = 3.63PTDD160 pKa = 3.95PPTDD164 pKa = 3.63PTDD167 pKa = 3.95PPTDD171 pKa = 3.63PTDD174 pKa = 3.81PPTDD178 pKa = 3.74PGEE181 pKa = 4.42RR182 pKa = 11.84VDD184 pKa = 4.09NPFVGAQGYY193 pKa = 7.83VNPVWSAKK201 pKa = 9.93AAAEE205 pKa = 4.17PGGSAVANEE214 pKa = 4.36STAVWLDD221 pKa = 3.49RR222 pKa = 11.84IGAIEE227 pKa = 4.59GNDD230 pKa = 3.48SATTGSMGLRR240 pKa = 11.84DD241 pKa = 4.31HH242 pKa = 7.21LDD244 pKa = 3.43EE245 pKa = 5.92AVAQADD251 pKa = 3.79GDD253 pKa = 4.1PLVIQVVIYY262 pKa = 9.28NLPGRR267 pKa = 11.84DD268 pKa = 3.5CAALASNGEE277 pKa = 4.27LGPEE281 pKa = 4.01EE282 pKa = 3.7IDD284 pKa = 3.36RR285 pKa = 11.84YY286 pKa = 9.4KK287 pKa = 11.04NEE289 pKa = 4.53YY290 pKa = 9.08IDD292 pKa = 5.08PIADD296 pKa = 3.08IMSDD300 pKa = 3.34YY301 pKa = 11.12AQYY304 pKa = 11.69DD305 pKa = 3.33NLRR308 pKa = 11.84IVNIIEE314 pKa = 4.22IDD316 pKa = 3.67SLPNLVTNTTDD327 pKa = 3.23NAGGTEE333 pKa = 4.44LCDD336 pKa = 3.56QMKK339 pKa = 11.14ANGNYY344 pKa = 9.64VNGVGYY350 pKa = 10.99ALATLGDD357 pKa = 3.65IPNAYY362 pKa = 10.11NYY364 pKa = 10.42VDD366 pKa = 3.89AAHH369 pKa = 6.77HH370 pKa = 6.27GWIGWDD376 pKa = 3.51TNFGPSAEE384 pKa = 4.2IFYY387 pKa = 10.41EE388 pKa = 4.09AANASGATVDD398 pKa = 4.47DD399 pKa = 3.59VHH401 pKa = 8.17GFIANTANYY410 pKa = 9.41SALEE414 pKa = 3.96EE415 pKa = 4.91PYY417 pKa = 10.82LDD419 pKa = 4.14VNGTVNGQMIRR430 pKa = 11.84QSDD433 pKa = 3.58WVDD436 pKa = 2.5WNQYY440 pKa = 8.86VDD442 pKa = 4.11EE443 pKa = 5.26LSFAQDD449 pKa = 3.18LRR451 pKa = 11.84QEE453 pKa = 4.15LVSVGFNSDD462 pKa = 2.43IGMLIDD468 pKa = 3.66TSRR471 pKa = 11.84NGWGGPEE478 pKa = 4.16RR479 pKa = 11.84PTGPSSATDD488 pKa = 3.47LNTYY492 pKa = 9.16VEE494 pKa = 4.42EE495 pKa = 4.13SRR497 pKa = 11.84IDD499 pKa = 3.37RR500 pKa = 11.84RR501 pKa = 11.84FNPGNWCNQAGAGLGEE517 pKa = 4.81RR518 pKa = 11.84PTAAPEE524 pKa = 4.12PGIDD528 pKa = 3.16AYY530 pKa = 11.33VWAKK534 pKa = 10.47PPGEE538 pKa = 3.93SDD540 pKa = 3.43GASEE544 pKa = 4.84YY545 pKa = 10.62IEE547 pKa = 4.38NDD549 pKa = 2.82EE550 pKa = 4.45GKK552 pKa = 10.7GFDD555 pKa = 4.06EE556 pKa = 4.87MCDD559 pKa = 3.29PAYY562 pKa = 10.57GGNARR567 pKa = 11.84NGNSPSGALPNAPISGHH584 pKa = 5.18WFSAQFQEE592 pKa = 4.77LLANAYY598 pKa = 8.53PPLL601 pKa = 5.25
MM1 pKa = 7.26SKK3 pKa = 10.02VRR5 pKa = 11.84ATNRR9 pKa = 11.84RR10 pKa = 11.84SWLRR14 pKa = 11.84RR15 pKa = 11.84GFATASSLALGASMVALAAPANAAGCSVDD44 pKa = 3.52YY45 pKa = 10.63QVNSWGSGFTASVTITNLGSALSNWTLEE73 pKa = 3.46WDD75 pKa = 4.02FPGNQQVTNLWNGSYY90 pKa = 9.81SQSGQHH96 pKa = 5.44VSVDD100 pKa = 3.3NAPYY104 pKa = 9.93NGSVGEE110 pKa = 4.25GGTVNFGFNGSYY122 pKa = 10.11SGSNDD127 pKa = 2.6IPTSFSLNGVTCDD140 pKa = 3.94GSVDD144 pKa = 4.01PDD146 pKa = 4.08PPTDD150 pKa = 3.7PTDD153 pKa = 3.95PPTDD157 pKa = 3.63PTDD160 pKa = 3.95PPTDD164 pKa = 3.63PTDD167 pKa = 3.95PPTDD171 pKa = 3.63PTDD174 pKa = 3.81PPTDD178 pKa = 3.74PGEE181 pKa = 4.42RR182 pKa = 11.84VDD184 pKa = 4.09NPFVGAQGYY193 pKa = 7.83VNPVWSAKK201 pKa = 9.93AAAEE205 pKa = 4.17PGGSAVANEE214 pKa = 4.36STAVWLDD221 pKa = 3.49RR222 pKa = 11.84IGAIEE227 pKa = 4.59GNDD230 pKa = 3.48SATTGSMGLRR240 pKa = 11.84DD241 pKa = 4.31HH242 pKa = 7.21LDD244 pKa = 3.43EE245 pKa = 5.92AVAQADD251 pKa = 3.79GDD253 pKa = 4.1PLVIQVVIYY262 pKa = 9.28NLPGRR267 pKa = 11.84DD268 pKa = 3.5CAALASNGEE277 pKa = 4.27LGPEE281 pKa = 4.01EE282 pKa = 3.7IDD284 pKa = 3.36RR285 pKa = 11.84YY286 pKa = 9.4KK287 pKa = 11.04NEE289 pKa = 4.53YY290 pKa = 9.08IDD292 pKa = 5.08PIADD296 pKa = 3.08IMSDD300 pKa = 3.34YY301 pKa = 11.12AQYY304 pKa = 11.69DD305 pKa = 3.33NLRR308 pKa = 11.84IVNIIEE314 pKa = 4.22IDD316 pKa = 3.67SLPNLVTNTTDD327 pKa = 3.23NAGGTEE333 pKa = 4.44LCDD336 pKa = 3.56QMKK339 pKa = 11.14ANGNYY344 pKa = 9.64VNGVGYY350 pKa = 10.99ALATLGDD357 pKa = 3.65IPNAYY362 pKa = 10.11NYY364 pKa = 10.42VDD366 pKa = 3.89AAHH369 pKa = 6.77HH370 pKa = 6.27GWIGWDD376 pKa = 3.51TNFGPSAEE384 pKa = 4.2IFYY387 pKa = 10.41EE388 pKa = 4.09AANASGATVDD398 pKa = 4.47DD399 pKa = 3.59VHH401 pKa = 8.17GFIANTANYY410 pKa = 9.41SALEE414 pKa = 3.96EE415 pKa = 4.91PYY417 pKa = 10.82LDD419 pKa = 4.14VNGTVNGQMIRR430 pKa = 11.84QSDD433 pKa = 3.58WVDD436 pKa = 2.5WNQYY440 pKa = 8.86VDD442 pKa = 4.11EE443 pKa = 5.26LSFAQDD449 pKa = 3.18LRR451 pKa = 11.84QEE453 pKa = 4.15LVSVGFNSDD462 pKa = 2.43IGMLIDD468 pKa = 3.66TSRR471 pKa = 11.84NGWGGPEE478 pKa = 4.16RR479 pKa = 11.84PTGPSSATDD488 pKa = 3.47LNTYY492 pKa = 9.16VEE494 pKa = 4.42EE495 pKa = 4.13SRR497 pKa = 11.84IDD499 pKa = 3.37RR500 pKa = 11.84RR501 pKa = 11.84FNPGNWCNQAGAGLGEE517 pKa = 4.81RR518 pKa = 11.84PTAAPEE524 pKa = 4.12PGIDD528 pKa = 3.16AYY530 pKa = 11.33VWAKK534 pKa = 10.47PPGEE538 pKa = 3.93SDD540 pKa = 3.43GASEE544 pKa = 4.84YY545 pKa = 10.62IEE547 pKa = 4.38NDD549 pKa = 2.82EE550 pKa = 4.45GKK552 pKa = 10.7GFDD555 pKa = 4.06EE556 pKa = 4.87MCDD559 pKa = 3.29PAYY562 pKa = 10.57GGNARR567 pKa = 11.84NGNSPSGALPNAPISGHH584 pKa = 5.18WFSAQFQEE592 pKa = 4.77LLANAYY598 pKa = 8.53PPLL601 pKa = 5.25
Molecular weight: 63.73 kDa
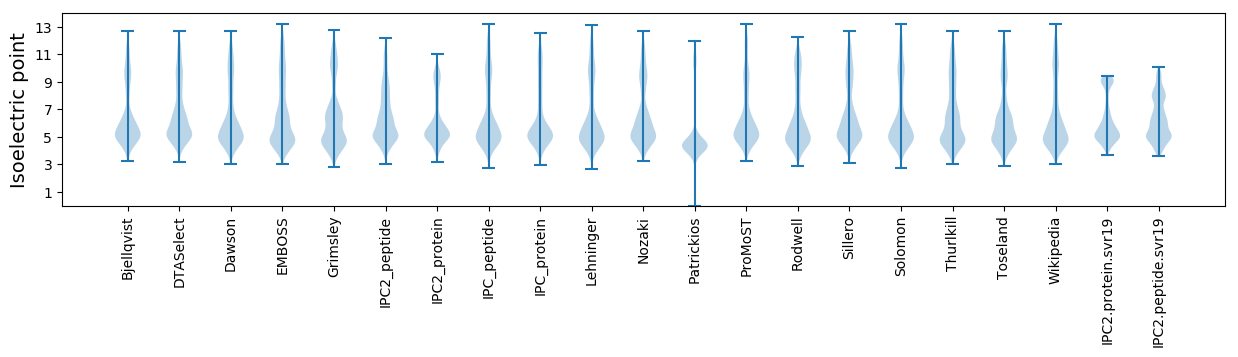
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A399G4A9|A0A399G4A9_9ACTN Cytochrome P450 OS=Thermobifida halotolerans OX=483545 GN=NI17_06900 PE=3 SV=1
MM1 pKa = 7.61RR2 pKa = 11.84FVPPSPARR10 pKa = 11.84ITPRR14 pKa = 11.84IRR16 pKa = 11.84LVTQTRR22 pKa = 11.84RR23 pKa = 11.84FVARR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 7.69TRR32 pKa = 11.84PRR34 pKa = 11.84PDD36 pKa = 3.0GGIRR40 pKa = 11.84PAAVGEE46 pKa = 3.81ARR48 pKa = 11.84YY49 pKa = 9.44RR50 pKa = 11.84CPGRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84SGRR59 pKa = 11.84TGRR62 pKa = 11.84RR63 pKa = 11.84AAPARR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84GPLPRR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84APRR81 pKa = 11.84LRR83 pKa = 11.84PGRR86 pKa = 11.84GRR88 pKa = 11.84TAA90 pKa = 3.11
MM1 pKa = 7.61RR2 pKa = 11.84FVPPSPARR10 pKa = 11.84ITPRR14 pKa = 11.84IRR16 pKa = 11.84LVTQTRR22 pKa = 11.84RR23 pKa = 11.84FVARR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 7.69TRR32 pKa = 11.84PRR34 pKa = 11.84PDD36 pKa = 3.0GGIRR40 pKa = 11.84PAAVGEE46 pKa = 3.81ARR48 pKa = 11.84YY49 pKa = 9.44RR50 pKa = 11.84CPGRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84SGRR59 pKa = 11.84TGRR62 pKa = 11.84RR63 pKa = 11.84AAPARR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84GPLPRR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84APRR81 pKa = 11.84LRR83 pKa = 11.84PGRR86 pKa = 11.84GRR88 pKa = 11.84TAA90 pKa = 3.11
Molecular weight: 10.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1241983 |
34 |
3574 |
305.4 |
32.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.751 ± 0.056 | 0.754 ± 0.011 |
6.285 ± 0.032 | 6.478 ± 0.035 |
2.809 ± 0.021 | 8.931 ± 0.041 |
2.304 ± 0.022 | 3.186 ± 0.023 |
1.604 ± 0.021 | 10.381 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.767 ± 0.015 | 1.859 ± 0.022 |
5.919 ± 0.026 | 2.588 ± 0.021 |
8.763 ± 0.044 | 5.071 ± 0.028 |
5.878 ± 0.033 | 9.097 ± 0.042 |
1.493 ± 0.018 | 2.083 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
