
Human endogenous retrovirus HCML-ARV
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; unclassified Retroviridae; Human endogenous retroviruses; Human endogenous retrovirus
Average proteome isoelectric point is 7.59
Get precalculated fractions of proteins
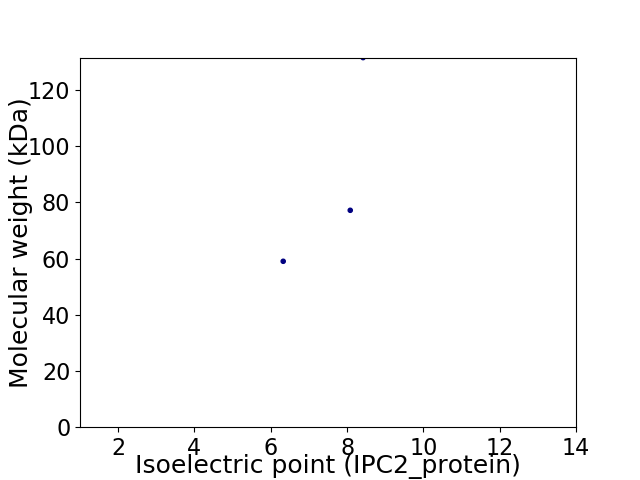
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q7ZGS4|Q7ZGS4_9RETR Retroviral gag protein OS=Human endogenous retrovirus HCML-ARV OX=228277 PE=4 SV=1
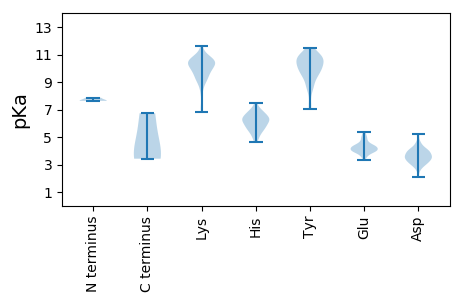
MM1 pKa = 7.63LKK3 pKa = 10.26NFKK6 pKa = 10.36KK7 pKa = 10.67GFNGDD12 pKa = 3.62YY13 pKa = 10.08GVTMTPGKK21 pKa = 10.46LRR23 pKa = 11.84ILCEE27 pKa = 4.13IDD29 pKa = 3.06WPTLEE34 pKa = 4.62VGWPSEE40 pKa = 3.91GSLDD44 pKa = 3.4RR45 pKa = 11.84SLVSKK50 pKa = 10.13VWHH53 pKa = 6.23KK54 pKa = 9.68VTGKK58 pKa = 10.11SGHH61 pKa = 6.23SDD63 pKa = 3.07QFPYY67 pKa = 10.4IDD69 pKa = 3.26TWLLQLVQDD78 pKa = 4.23PPQWLRR84 pKa = 11.84GQAAAVLVAKK94 pKa = 10.01GQIAKK99 pKa = 9.83EE100 pKa = 4.55GSRR103 pKa = 11.84STHH106 pKa = 5.62WGKK109 pKa = 8.47STPEE113 pKa = 3.79VLFDD117 pKa = 3.9PTSEE121 pKa = 5.11DD122 pKa = 3.82PLQEE126 pKa = 4.18MAPVIPVLPSPYY138 pKa = 9.24QAEE141 pKa = 4.03RR142 pKa = 11.84LPTFEE147 pKa = 4.49PTVLVPPQDD156 pKa = 3.16KK157 pKa = 10.56HH158 pKa = 7.02IPRR161 pKa = 11.84PPRR164 pKa = 11.84VDD166 pKa = 2.92KK167 pKa = 11.27RR168 pKa = 11.84GGEE171 pKa = 4.17ASGEE175 pKa = 4.3TPPLAACLRR184 pKa = 11.84PKK186 pKa = 10.23TGIQMPLRR194 pKa = 11.84EE195 pKa = 3.95QRR197 pKa = 11.84YY198 pKa = 7.07TGIEE202 pKa = 3.97EE203 pKa = 4.52DD204 pKa = 3.11GHH206 pKa = 5.06MVEE209 pKa = 5.01KK210 pKa = 10.43RR211 pKa = 11.84VFVYY215 pKa = 10.58QPFTSANLLNWKK227 pKa = 9.98NNTLSYY233 pKa = 9.95TEE235 pKa = 4.87KK236 pKa = 10.24PQALIDD242 pKa = 4.01LLQTIIQTHH251 pKa = 5.78NSTRR255 pKa = 11.84ADD257 pKa = 3.52CHH259 pKa = 5.93QLLMFLFNTDD269 pKa = 2.09EE270 pKa = 4.22RR271 pKa = 11.84QRR273 pKa = 11.84VLQAATKK280 pKa = 8.79WVQEE284 pKa = 4.27HH285 pKa = 6.82APADD289 pKa = 3.69YY290 pKa = 10.76QNPQEE295 pKa = 4.42CVRR298 pKa = 11.84TQLPGTDD305 pKa = 4.01PQWDD309 pKa = 3.67PNEE312 pKa = 4.31RR313 pKa = 11.84EE314 pKa = 3.87DD315 pKa = 3.7MQRR318 pKa = 11.84LNRR321 pKa = 11.84DD322 pKa = 3.0RR323 pKa = 11.84EE324 pKa = 4.26AVLEE328 pKa = 3.97GLKK331 pKa = 10.55RR332 pKa = 11.84GAQKK336 pKa = 9.47ATNVNKK342 pKa = 9.77VSEE345 pKa = 4.46VIRR348 pKa = 11.84GKK350 pKa = 10.51EE351 pKa = 3.54EE352 pKa = 4.0SPAQFYY358 pKa = 10.81QRR360 pKa = 11.84LCEE363 pKa = 4.34GYY365 pKa = 10.82RR366 pKa = 11.84MYY368 pKa = 11.01TPFDD372 pKa = 3.52PVSPEE377 pKa = 3.68NQRR380 pKa = 11.84MVNMALVSQSAEE392 pKa = 4.01DD393 pKa = 3.97IRR395 pKa = 11.84RR396 pKa = 11.84KK397 pKa = 9.24LQKK400 pKa = 9.73QDD402 pKa = 3.01GFAGTNTSQLLEE414 pKa = 4.0VANQVFVNRR423 pKa = 11.84DD424 pKa = 3.02AVSPKK429 pKa = 9.56EE430 pKa = 3.75NRR432 pKa = 11.84RR433 pKa = 11.84EE434 pKa = 3.87NEE436 pKa = 3.47RR437 pKa = 11.84QARR440 pKa = 11.84RR441 pKa = 11.84NAEE444 pKa = 3.96LLAAAVGGVSSKK456 pKa = 10.68RR457 pKa = 11.84QGKK460 pKa = 9.35GGPGKK465 pKa = 8.6EE466 pKa = 4.23TQPGCQSLQCNQCAYY481 pKa = 10.08CKK483 pKa = 10.51EE484 pKa = 3.37IGYY487 pKa = 8.55WKK489 pKa = 10.42NKK491 pKa = 9.25CPQLKK496 pKa = 10.08GKK498 pKa = 10.18QGDD501 pKa = 4.11LEE503 pKa = 4.37QEE505 pKa = 4.3VPDD508 pKa = 4.24KK509 pKa = 11.65EE510 pKa = 4.2EE511 pKa = 4.06GALLNLAEE519 pKa = 4.74EE520 pKa = 4.63LLDD523 pKa = 3.77
MM1 pKa = 7.63LKK3 pKa = 10.26NFKK6 pKa = 10.36KK7 pKa = 10.67GFNGDD12 pKa = 3.62YY13 pKa = 10.08GVTMTPGKK21 pKa = 10.46LRR23 pKa = 11.84ILCEE27 pKa = 4.13IDD29 pKa = 3.06WPTLEE34 pKa = 4.62VGWPSEE40 pKa = 3.91GSLDD44 pKa = 3.4RR45 pKa = 11.84SLVSKK50 pKa = 10.13VWHH53 pKa = 6.23KK54 pKa = 9.68VTGKK58 pKa = 10.11SGHH61 pKa = 6.23SDD63 pKa = 3.07QFPYY67 pKa = 10.4IDD69 pKa = 3.26TWLLQLVQDD78 pKa = 4.23PPQWLRR84 pKa = 11.84GQAAAVLVAKK94 pKa = 10.01GQIAKK99 pKa = 9.83EE100 pKa = 4.55GSRR103 pKa = 11.84STHH106 pKa = 5.62WGKK109 pKa = 8.47STPEE113 pKa = 3.79VLFDD117 pKa = 3.9PTSEE121 pKa = 5.11DD122 pKa = 3.82PLQEE126 pKa = 4.18MAPVIPVLPSPYY138 pKa = 9.24QAEE141 pKa = 4.03RR142 pKa = 11.84LPTFEE147 pKa = 4.49PTVLVPPQDD156 pKa = 3.16KK157 pKa = 10.56HH158 pKa = 7.02IPRR161 pKa = 11.84PPRR164 pKa = 11.84VDD166 pKa = 2.92KK167 pKa = 11.27RR168 pKa = 11.84GGEE171 pKa = 4.17ASGEE175 pKa = 4.3TPPLAACLRR184 pKa = 11.84PKK186 pKa = 10.23TGIQMPLRR194 pKa = 11.84EE195 pKa = 3.95QRR197 pKa = 11.84YY198 pKa = 7.07TGIEE202 pKa = 3.97EE203 pKa = 4.52DD204 pKa = 3.11GHH206 pKa = 5.06MVEE209 pKa = 5.01KK210 pKa = 10.43RR211 pKa = 11.84VFVYY215 pKa = 10.58QPFTSANLLNWKK227 pKa = 9.98NNTLSYY233 pKa = 9.95TEE235 pKa = 4.87KK236 pKa = 10.24PQALIDD242 pKa = 4.01LLQTIIQTHH251 pKa = 5.78NSTRR255 pKa = 11.84ADD257 pKa = 3.52CHH259 pKa = 5.93QLLMFLFNTDD269 pKa = 2.09EE270 pKa = 4.22RR271 pKa = 11.84QRR273 pKa = 11.84VLQAATKK280 pKa = 8.79WVQEE284 pKa = 4.27HH285 pKa = 6.82APADD289 pKa = 3.69YY290 pKa = 10.76QNPQEE295 pKa = 4.42CVRR298 pKa = 11.84TQLPGTDD305 pKa = 4.01PQWDD309 pKa = 3.67PNEE312 pKa = 4.31RR313 pKa = 11.84EE314 pKa = 3.87DD315 pKa = 3.7MQRR318 pKa = 11.84LNRR321 pKa = 11.84DD322 pKa = 3.0RR323 pKa = 11.84EE324 pKa = 4.26AVLEE328 pKa = 3.97GLKK331 pKa = 10.55RR332 pKa = 11.84GAQKK336 pKa = 9.47ATNVNKK342 pKa = 9.77VSEE345 pKa = 4.46VIRR348 pKa = 11.84GKK350 pKa = 10.51EE351 pKa = 3.54EE352 pKa = 4.0SPAQFYY358 pKa = 10.81QRR360 pKa = 11.84LCEE363 pKa = 4.34GYY365 pKa = 10.82RR366 pKa = 11.84MYY368 pKa = 11.01TPFDD372 pKa = 3.52PVSPEE377 pKa = 3.68NQRR380 pKa = 11.84MVNMALVSQSAEE392 pKa = 4.01DD393 pKa = 3.97IRR395 pKa = 11.84RR396 pKa = 11.84KK397 pKa = 9.24LQKK400 pKa = 9.73QDD402 pKa = 3.01GFAGTNTSQLLEE414 pKa = 4.0VANQVFVNRR423 pKa = 11.84DD424 pKa = 3.02AVSPKK429 pKa = 9.56EE430 pKa = 3.75NRR432 pKa = 11.84RR433 pKa = 11.84EE434 pKa = 3.87NEE436 pKa = 3.47RR437 pKa = 11.84QARR440 pKa = 11.84RR441 pKa = 11.84NAEE444 pKa = 3.96LLAAAVGGVSSKK456 pKa = 10.68RR457 pKa = 11.84QGKK460 pKa = 9.35GGPGKK465 pKa = 8.6EE466 pKa = 4.23TQPGCQSLQCNQCAYY481 pKa = 10.08CKK483 pKa = 10.51EE484 pKa = 3.37IGYY487 pKa = 8.55WKK489 pKa = 10.42NKK491 pKa = 9.25CPQLKK496 pKa = 10.08GKK498 pKa = 10.18QGDD501 pKa = 4.11LEE503 pKa = 4.37QEE505 pKa = 4.3VPDD508 pKa = 4.24KK509 pKa = 11.65EE510 pKa = 4.2EE511 pKa = 4.06GALLNLAEE519 pKa = 4.74EE520 pKa = 4.63LLDD523 pKa = 3.77
Molecular weight: 59.02 kDa
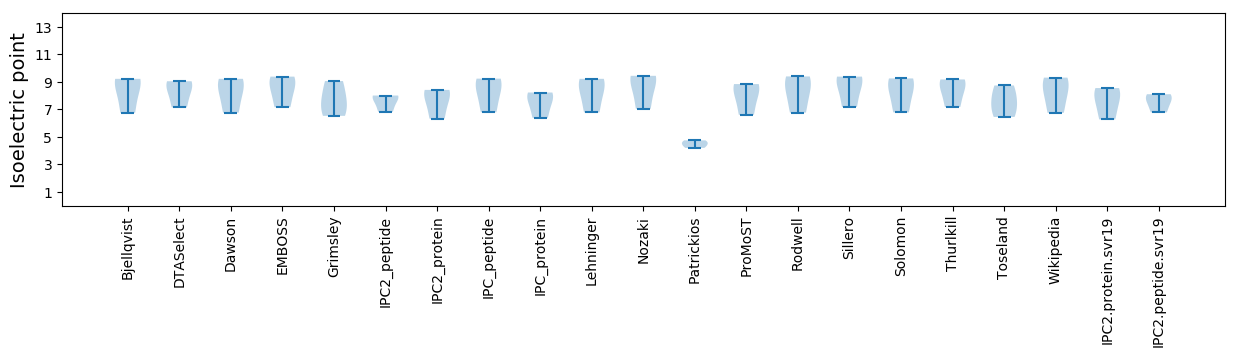
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q7ZGS4|Q7ZGS4_9RETR Retroviral gag protein OS=Human endogenous retrovirus HCML-ARV OX=228277 PE=4 SV=1
MM1 pKa = 7.63ARR3 pKa = 11.84MTVGGKK9 pKa = 10.16DD10 pKa = 2.84IDD12 pKa = 4.04FLVDD16 pKa = 3.28TGAEE20 pKa = 4.15HH21 pKa = 6.87SVVTAPVAPLSKK33 pKa = 9.73KK34 pKa = 9.21TIDD37 pKa = 3.59IIGAMGVSAKK47 pKa = 9.95QAFCLPRR54 pKa = 11.84TCTVGGHH61 pKa = 5.09KK62 pKa = 10.27VIHH65 pKa = 5.97QFLYY69 pKa = 10.35MPDD72 pKa = 3.85CPLPLLGRR80 pKa = 11.84DD81 pKa = 3.68LLSKK85 pKa = 10.69LRR87 pKa = 11.84ATISFTEE94 pKa = 4.27HH95 pKa = 6.48GSLLLKK101 pKa = 10.92LPGTGVIMTLTVPRR115 pKa = 11.84EE116 pKa = 4.06EE117 pKa = 4.03EE118 pKa = 3.47WRR120 pKa = 11.84LFLTEE125 pKa = 4.16SGQEE129 pKa = 3.76IRR131 pKa = 11.84PALAKK136 pKa = 9.96RR137 pKa = 11.84WPRR140 pKa = 11.84VWAEE144 pKa = 4.13DD145 pKa = 3.97NPPGLAVNQAPVLIEE160 pKa = 4.16VKK162 pKa = 10.47PGAQPVRR169 pKa = 11.84QKK171 pKa = 10.57QYY173 pKa = 9.4PVPRR177 pKa = 11.84EE178 pKa = 3.98ALEE181 pKa = 4.94GIQVPLKK188 pKa = 10.17CLRR191 pKa = 11.84TFGMIVPCQSPWNTPLLPVPEE212 pKa = 5.37PKK214 pKa = 10.23TKK216 pKa = 10.41DD217 pKa = 3.24YY218 pKa = 11.13WPGQDD223 pKa = 4.26LRR225 pKa = 11.84LVKK228 pKa = 10.3QATVTLHH235 pKa = 6.24PAVPNPYY242 pKa = 8.66TLLGLLPAEE251 pKa = 4.96DD252 pKa = 3.94SCFTCLDD259 pKa = 3.54LKK261 pKa = 11.19DD262 pKa = 3.98AFFSIRR268 pKa = 11.84FAPEE272 pKa = 3.69SQKK275 pKa = 10.98LFAFQWEE282 pKa = 4.4DD283 pKa = 3.47PEE285 pKa = 5.34SGVTTQYY292 pKa = 9.45TWTWLPQGFKK302 pKa = 10.67NSPTIFGEE310 pKa = 3.92EE311 pKa = 4.24LARR314 pKa = 11.84DD315 pKa = 3.78LQKK318 pKa = 11.19CPTRR322 pKa = 11.84DD323 pKa = 3.47LGCVLLQYY331 pKa = 11.08VDD333 pKa = 5.2DD334 pKa = 5.23LLLGHH339 pKa = 6.79PTAVGCAKK347 pKa = 9.79GTDD350 pKa = 3.06ALLRR354 pKa = 11.84HH355 pKa = 6.45LEE357 pKa = 4.13DD358 pKa = 3.96CGYY361 pKa = 10.72KK362 pKa = 10.05VSKK365 pKa = 10.39KK366 pKa = 9.79KK367 pKa = 10.77AQICRR372 pKa = 11.84PQVRR376 pKa = 11.84YY377 pKa = 10.19LGFTIRR383 pKa = 11.84QGEE386 pKa = 4.05RR387 pKa = 11.84SLGSEE392 pKa = 4.54RR393 pKa = 11.84KK394 pKa = 8.9QVICTLPEE402 pKa = 4.11PKK404 pKa = 9.78SRR406 pKa = 11.84KK407 pKa = 6.87QVRR410 pKa = 11.84KK411 pKa = 9.13FLGAAGFCRR420 pKa = 11.84LWIPNFAVLAKK431 pKa = 9.96PLYY434 pKa = 10.18EE435 pKa = 3.94VTKK438 pKa = 10.04WGDD441 pKa = 3.31RR442 pKa = 11.84EE443 pKa = 4.01PFEE446 pKa = 4.96WGSQQQQAFRR456 pKa = 11.84EE457 pKa = 4.34LKK459 pKa = 9.44EE460 pKa = 4.27KK461 pKa = 10.77LMSAPALGLPNLTKK475 pKa = 10.63PFTLYY480 pKa = 10.2VSEE483 pKa = 4.24RR484 pKa = 11.84EE485 pKa = 4.05KK486 pKa = 10.46MAVRR490 pKa = 11.84VLTQTVGPWPRR501 pKa = 11.84PVAYY505 pKa = 10.33LSKK508 pKa = 10.82QLDD511 pKa = 4.2GVSKK515 pKa = 10.42GWPPCLRR522 pKa = 11.84ALAATALLVQEE533 pKa = 4.5AVKK536 pKa = 9.29LTLGQNLNIKK546 pKa = 9.68APHH549 pKa = 6.32AMVTLINTKK558 pKa = 8.1GHH560 pKa = 6.06HH561 pKa = 6.14WLTNARR567 pKa = 11.84LTKK570 pKa = 10.08YY571 pKa = 10.49QSLLCEE577 pKa = 4.23NPRR580 pKa = 11.84ITIEE584 pKa = 3.6ICNTLNPTTLLLVSEE599 pKa = 5.29GPVEE603 pKa = 4.12HH604 pKa = 7.47DD605 pKa = 3.45CVEE608 pKa = 4.29VLDD611 pKa = 4.29SVYY614 pKa = 10.96SSRR617 pKa = 11.84PDD619 pKa = 3.86LQDD622 pKa = 3.45QPWAPVDD629 pKa = 3.15WEE631 pKa = 4.66LYY633 pKa = 8.59MDD635 pKa = 4.7GGSFINPQGEE645 pKa = 4.03RR646 pKa = 11.84GAGYY650 pKa = 10.73AVVTLDD656 pKa = 3.38TVVEE660 pKa = 4.15ARR662 pKa = 11.84SLPQATSAQKK672 pKa = 10.89AEE674 pKa = 4.68LIAFIRR680 pKa = 11.84ALEE683 pKa = 4.06LSEE686 pKa = 4.83GEE688 pKa = 4.43TVNIYY693 pKa = 10.01TDD695 pKa = 2.81SRR697 pKa = 11.84YY698 pKa = 9.87PFLTLQVHH706 pKa = 5.21EE707 pKa = 4.79ASYY710 pKa = 10.98KK711 pKa = 10.61EE712 pKa = 3.97KK713 pKa = 11.0GLLNSGGKK721 pKa = 9.51DD722 pKa = 2.53IKK724 pKa = 10.48YY725 pKa = 9.12QQEE728 pKa = 3.77ILQLLEE734 pKa = 4.96AVWKK738 pKa = 7.04PHH740 pKa = 5.46KK741 pKa = 10.21VAVMHH746 pKa = 6.47CRR748 pKa = 11.84GHH750 pKa = 5.39QRR752 pKa = 11.84ASTLVGLGNSRR763 pKa = 11.84ADD765 pKa = 3.32ARR767 pKa = 11.84KK768 pKa = 9.04AASAPFRR775 pKa = 11.84ASVTAPLLPQAPDD788 pKa = 3.73LLPTYY793 pKa = 10.67SKK795 pKa = 11.0EE796 pKa = 4.3EE797 pKa = 3.94KK798 pKa = 10.48DD799 pKa = 3.53FLQAEE804 pKa = 5.33GGQVMEE810 pKa = 5.01EE811 pKa = 3.95GWIRR815 pKa = 11.84LPDD818 pKa = 3.49GRR820 pKa = 11.84EE821 pKa = 3.79AVPQLLGAAVVLAVHH836 pKa = 6.4KK837 pKa = 6.85TTHH840 pKa = 6.85LGQEE844 pKa = 4.32SLEE847 pKa = 4.21KK848 pKa = 10.87LLVRR852 pKa = 11.84YY853 pKa = 8.39FYY855 pKa = 10.5ILHH858 pKa = 6.75LSALAKK864 pKa = 9.76TVTQRR869 pKa = 11.84CVTCPKK875 pKa = 10.25HH876 pKa = 5.98NAKK879 pKa = 10.08QGPAVPPVIQAYY891 pKa = 9.37GAAPFEE897 pKa = 4.37DD898 pKa = 3.74VQVDD902 pKa = 4.04FTEE905 pKa = 4.31MPKK908 pKa = 10.71CGGNKK913 pKa = 9.18YY914 pKa = 10.7LLVLVCTYY922 pKa = 10.9SGGWRR927 pKa = 11.84LIQHH931 pKa = 7.17KK932 pKa = 9.93LRR934 pKa = 11.84KK935 pKa = 8.92LVKK938 pKa = 8.68LTCVLLRR945 pKa = 11.84DD946 pKa = 4.64PIPRR950 pKa = 11.84FGLPLRR956 pKa = 11.84IGSDD960 pKa = 3.05NGPAFVADD968 pKa = 4.47LVQKK972 pKa = 10.03RR973 pKa = 11.84AKK975 pKa = 10.59VLGNTWKK982 pKa = 10.35LHH984 pKa = 4.82AAYY987 pKa = 10.0RR988 pKa = 11.84PQSSGKK994 pKa = 9.05VEE996 pKa = 3.87QMNWTIKK1003 pKa = 9.43NSKK1006 pKa = 10.34GKK1008 pKa = 9.72VCQEE1012 pKa = 4.39TGLKK1016 pKa = 9.21WIQALPMVLFKK1027 pKa = 10.57IRR1029 pKa = 11.84CTPSKK1034 pKa = 9.46RR1035 pKa = 11.84TGYY1038 pKa = 10.04SPYY1041 pKa = 9.84EE1042 pKa = 4.04ILYY1045 pKa = 9.54HH1046 pKa = 6.5RR1047 pKa = 11.84PPPILRR1053 pKa = 11.84GLPGTPRR1060 pKa = 11.84EE1061 pKa = 4.07LGEE1064 pKa = 5.09IEE1066 pKa = 5.17LQRR1069 pKa = 11.84QLQALGKK1076 pKa = 8.06ITQTISAWVNEE1087 pKa = 4.32RR1088 pKa = 11.84CPVSLFSPVHH1098 pKa = 5.89PFSPGNRR1105 pKa = 11.84VWIKK1109 pKa = 9.86DD1110 pKa = 3.33WNVASLCPRR1119 pKa = 11.84WKK1121 pKa = 10.69GPQTVILTTPTAVKK1135 pKa = 10.8VEE1137 pKa = 4.4GVPAWIHH1144 pKa = 5.39HH1145 pKa = 5.76SRR1147 pKa = 11.84VKK1149 pKa = 10.31PAVPEE1154 pKa = 4.03TSEE1157 pKa = 4.0VRR1159 pKa = 11.84PSPEE1163 pKa = 3.84DD1164 pKa = 3.32PCRR1167 pKa = 11.84VTLKK1171 pKa = 9.54KK1172 pKa = 7.36TTSPAPVTPGSS1183 pKa = 3.42
MM1 pKa = 7.63ARR3 pKa = 11.84MTVGGKK9 pKa = 10.16DD10 pKa = 2.84IDD12 pKa = 4.04FLVDD16 pKa = 3.28TGAEE20 pKa = 4.15HH21 pKa = 6.87SVVTAPVAPLSKK33 pKa = 9.73KK34 pKa = 9.21TIDD37 pKa = 3.59IIGAMGVSAKK47 pKa = 9.95QAFCLPRR54 pKa = 11.84TCTVGGHH61 pKa = 5.09KK62 pKa = 10.27VIHH65 pKa = 5.97QFLYY69 pKa = 10.35MPDD72 pKa = 3.85CPLPLLGRR80 pKa = 11.84DD81 pKa = 3.68LLSKK85 pKa = 10.69LRR87 pKa = 11.84ATISFTEE94 pKa = 4.27HH95 pKa = 6.48GSLLLKK101 pKa = 10.92LPGTGVIMTLTVPRR115 pKa = 11.84EE116 pKa = 4.06EE117 pKa = 4.03EE118 pKa = 3.47WRR120 pKa = 11.84LFLTEE125 pKa = 4.16SGQEE129 pKa = 3.76IRR131 pKa = 11.84PALAKK136 pKa = 9.96RR137 pKa = 11.84WPRR140 pKa = 11.84VWAEE144 pKa = 4.13DD145 pKa = 3.97NPPGLAVNQAPVLIEE160 pKa = 4.16VKK162 pKa = 10.47PGAQPVRR169 pKa = 11.84QKK171 pKa = 10.57QYY173 pKa = 9.4PVPRR177 pKa = 11.84EE178 pKa = 3.98ALEE181 pKa = 4.94GIQVPLKK188 pKa = 10.17CLRR191 pKa = 11.84TFGMIVPCQSPWNTPLLPVPEE212 pKa = 5.37PKK214 pKa = 10.23TKK216 pKa = 10.41DD217 pKa = 3.24YY218 pKa = 11.13WPGQDD223 pKa = 4.26LRR225 pKa = 11.84LVKK228 pKa = 10.3QATVTLHH235 pKa = 6.24PAVPNPYY242 pKa = 8.66TLLGLLPAEE251 pKa = 4.96DD252 pKa = 3.94SCFTCLDD259 pKa = 3.54LKK261 pKa = 11.19DD262 pKa = 3.98AFFSIRR268 pKa = 11.84FAPEE272 pKa = 3.69SQKK275 pKa = 10.98LFAFQWEE282 pKa = 4.4DD283 pKa = 3.47PEE285 pKa = 5.34SGVTTQYY292 pKa = 9.45TWTWLPQGFKK302 pKa = 10.67NSPTIFGEE310 pKa = 3.92EE311 pKa = 4.24LARR314 pKa = 11.84DD315 pKa = 3.78LQKK318 pKa = 11.19CPTRR322 pKa = 11.84DD323 pKa = 3.47LGCVLLQYY331 pKa = 11.08VDD333 pKa = 5.2DD334 pKa = 5.23LLLGHH339 pKa = 6.79PTAVGCAKK347 pKa = 9.79GTDD350 pKa = 3.06ALLRR354 pKa = 11.84HH355 pKa = 6.45LEE357 pKa = 4.13DD358 pKa = 3.96CGYY361 pKa = 10.72KK362 pKa = 10.05VSKK365 pKa = 10.39KK366 pKa = 9.79KK367 pKa = 10.77AQICRR372 pKa = 11.84PQVRR376 pKa = 11.84YY377 pKa = 10.19LGFTIRR383 pKa = 11.84QGEE386 pKa = 4.05RR387 pKa = 11.84SLGSEE392 pKa = 4.54RR393 pKa = 11.84KK394 pKa = 8.9QVICTLPEE402 pKa = 4.11PKK404 pKa = 9.78SRR406 pKa = 11.84KK407 pKa = 6.87QVRR410 pKa = 11.84KK411 pKa = 9.13FLGAAGFCRR420 pKa = 11.84LWIPNFAVLAKK431 pKa = 9.96PLYY434 pKa = 10.18EE435 pKa = 3.94VTKK438 pKa = 10.04WGDD441 pKa = 3.31RR442 pKa = 11.84EE443 pKa = 4.01PFEE446 pKa = 4.96WGSQQQQAFRR456 pKa = 11.84EE457 pKa = 4.34LKK459 pKa = 9.44EE460 pKa = 4.27KK461 pKa = 10.77LMSAPALGLPNLTKK475 pKa = 10.63PFTLYY480 pKa = 10.2VSEE483 pKa = 4.24RR484 pKa = 11.84EE485 pKa = 4.05KK486 pKa = 10.46MAVRR490 pKa = 11.84VLTQTVGPWPRR501 pKa = 11.84PVAYY505 pKa = 10.33LSKK508 pKa = 10.82QLDD511 pKa = 4.2GVSKK515 pKa = 10.42GWPPCLRR522 pKa = 11.84ALAATALLVQEE533 pKa = 4.5AVKK536 pKa = 9.29LTLGQNLNIKK546 pKa = 9.68APHH549 pKa = 6.32AMVTLINTKK558 pKa = 8.1GHH560 pKa = 6.06HH561 pKa = 6.14WLTNARR567 pKa = 11.84LTKK570 pKa = 10.08YY571 pKa = 10.49QSLLCEE577 pKa = 4.23NPRR580 pKa = 11.84ITIEE584 pKa = 3.6ICNTLNPTTLLLVSEE599 pKa = 5.29GPVEE603 pKa = 4.12HH604 pKa = 7.47DD605 pKa = 3.45CVEE608 pKa = 4.29VLDD611 pKa = 4.29SVYY614 pKa = 10.96SSRR617 pKa = 11.84PDD619 pKa = 3.86LQDD622 pKa = 3.45QPWAPVDD629 pKa = 3.15WEE631 pKa = 4.66LYY633 pKa = 8.59MDD635 pKa = 4.7GGSFINPQGEE645 pKa = 4.03RR646 pKa = 11.84GAGYY650 pKa = 10.73AVVTLDD656 pKa = 3.38TVVEE660 pKa = 4.15ARR662 pKa = 11.84SLPQATSAQKK672 pKa = 10.89AEE674 pKa = 4.68LIAFIRR680 pKa = 11.84ALEE683 pKa = 4.06LSEE686 pKa = 4.83GEE688 pKa = 4.43TVNIYY693 pKa = 10.01TDD695 pKa = 2.81SRR697 pKa = 11.84YY698 pKa = 9.87PFLTLQVHH706 pKa = 5.21EE707 pKa = 4.79ASYY710 pKa = 10.98KK711 pKa = 10.61EE712 pKa = 3.97KK713 pKa = 11.0GLLNSGGKK721 pKa = 9.51DD722 pKa = 2.53IKK724 pKa = 10.48YY725 pKa = 9.12QQEE728 pKa = 3.77ILQLLEE734 pKa = 4.96AVWKK738 pKa = 7.04PHH740 pKa = 5.46KK741 pKa = 10.21VAVMHH746 pKa = 6.47CRR748 pKa = 11.84GHH750 pKa = 5.39QRR752 pKa = 11.84ASTLVGLGNSRR763 pKa = 11.84ADD765 pKa = 3.32ARR767 pKa = 11.84KK768 pKa = 9.04AASAPFRR775 pKa = 11.84ASVTAPLLPQAPDD788 pKa = 3.73LLPTYY793 pKa = 10.67SKK795 pKa = 11.0EE796 pKa = 4.3EE797 pKa = 3.94KK798 pKa = 10.48DD799 pKa = 3.53FLQAEE804 pKa = 5.33GGQVMEE810 pKa = 5.01EE811 pKa = 3.95GWIRR815 pKa = 11.84LPDD818 pKa = 3.49GRR820 pKa = 11.84EE821 pKa = 3.79AVPQLLGAAVVLAVHH836 pKa = 6.4KK837 pKa = 6.85TTHH840 pKa = 6.85LGQEE844 pKa = 4.32SLEE847 pKa = 4.21KK848 pKa = 10.87LLVRR852 pKa = 11.84YY853 pKa = 8.39FYY855 pKa = 10.5ILHH858 pKa = 6.75LSALAKK864 pKa = 9.76TVTQRR869 pKa = 11.84CVTCPKK875 pKa = 10.25HH876 pKa = 5.98NAKK879 pKa = 10.08QGPAVPPVIQAYY891 pKa = 9.37GAAPFEE897 pKa = 4.37DD898 pKa = 3.74VQVDD902 pKa = 4.04FTEE905 pKa = 4.31MPKK908 pKa = 10.71CGGNKK913 pKa = 9.18YY914 pKa = 10.7LLVLVCTYY922 pKa = 10.9SGGWRR927 pKa = 11.84LIQHH931 pKa = 7.17KK932 pKa = 9.93LRR934 pKa = 11.84KK935 pKa = 8.92LVKK938 pKa = 8.68LTCVLLRR945 pKa = 11.84DD946 pKa = 4.64PIPRR950 pKa = 11.84FGLPLRR956 pKa = 11.84IGSDD960 pKa = 3.05NGPAFVADD968 pKa = 4.47LVQKK972 pKa = 10.03RR973 pKa = 11.84AKK975 pKa = 10.59VLGNTWKK982 pKa = 10.35LHH984 pKa = 4.82AAYY987 pKa = 10.0RR988 pKa = 11.84PQSSGKK994 pKa = 9.05VEE996 pKa = 3.87QMNWTIKK1003 pKa = 9.43NSKK1006 pKa = 10.34GKK1008 pKa = 9.72VCQEE1012 pKa = 4.39TGLKK1016 pKa = 9.21WIQALPMVLFKK1027 pKa = 10.57IRR1029 pKa = 11.84CTPSKK1034 pKa = 9.46RR1035 pKa = 11.84TGYY1038 pKa = 10.04SPYY1041 pKa = 9.84EE1042 pKa = 4.04ILYY1045 pKa = 9.54HH1046 pKa = 6.5RR1047 pKa = 11.84PPPILRR1053 pKa = 11.84GLPGTPRR1060 pKa = 11.84EE1061 pKa = 4.07LGEE1064 pKa = 5.09IEE1066 pKa = 5.17LQRR1069 pKa = 11.84QLQALGKK1076 pKa = 8.06ITQTISAWVNEE1087 pKa = 4.32RR1088 pKa = 11.84CPVSLFSPVHH1098 pKa = 5.89PFSPGNRR1105 pKa = 11.84VWIKK1109 pKa = 9.86DD1110 pKa = 3.33WNVASLCPRR1119 pKa = 11.84WKK1121 pKa = 10.69GPQTVILTTPTAVKK1135 pKa = 10.8VEE1137 pKa = 4.4GVPAWIHH1144 pKa = 5.39HH1145 pKa = 5.76SRR1147 pKa = 11.84VKK1149 pKa = 10.31PAVPEE1154 pKa = 4.03TSEE1157 pKa = 4.0VRR1159 pKa = 11.84PSPEE1163 pKa = 3.84DD1164 pKa = 3.32PCRR1167 pKa = 11.84VTLKK1171 pKa = 9.54KK1172 pKa = 7.36TTSPAPVTPGSS1183 pKa = 3.42
Molecular weight: 131.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2388 |
523 |
1183 |
796.0 |
89.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.658 ± 0.612 | 2.596 ± 0.334 |
3.727 ± 0.403 | 6.03 ± 0.656 |
3.015 ± 0.275 | 6.826 ± 0.039 |
2.052 ± 0.179 | 4.523 ± 1.097 |
6.407 ± 0.032 | 10.469 ± 0.849 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.34 ± 0.195 | 3.601 ± 0.834 |
7.58 ± 0.79 | 5.695 ± 0.752 |
5.486 ± 0.431 | 4.899 ± 0.044 |
6.826 ± 0.675 | 7.161 ± 0.492 |
2.387 ± 0.234 | 2.722 ± 0.322 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
