
Changjiang tombus-like virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.4
Get precalculated fractions of proteins
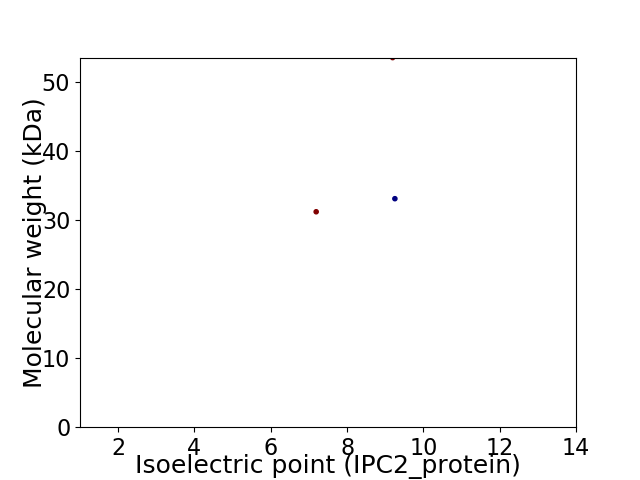
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
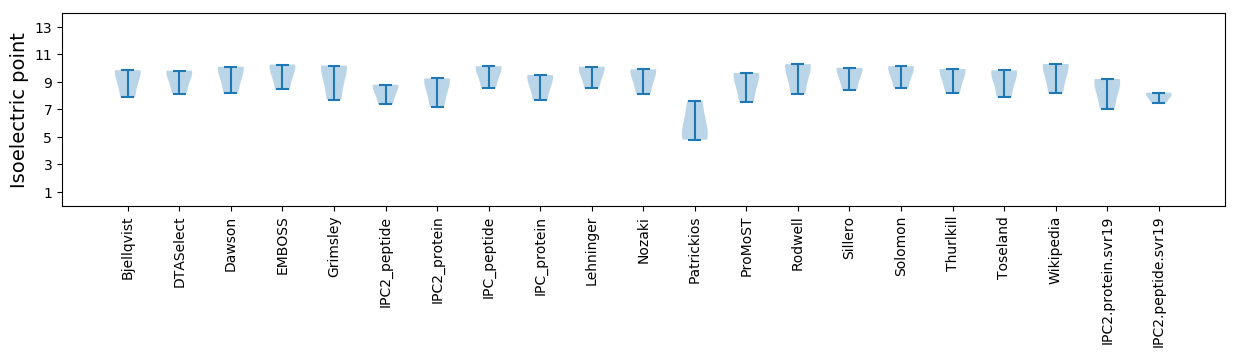
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFW2|A0A1L3KFW2_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 4 OX=1922818 PE=4 SV=1
MM1 pKa = 7.07IQVPSRR7 pKa = 11.84QGLQAIGGGSTEE19 pKa = 3.83QLRR22 pKa = 11.84SHH24 pKa = 7.04RR25 pKa = 11.84PTLLGLFEE33 pKa = 5.74DD34 pKa = 4.33GWMGYY39 pKa = 9.49PSALSPSTAGDD50 pKa = 3.57VEE52 pKa = 4.66VPKK55 pKa = 10.83AQAEE59 pKa = 4.6VPSNNARR66 pKa = 11.84GPLFGLGEE74 pKa = 4.43SPTAKK79 pKa = 10.0TPGEE83 pKa = 4.35TPFSQVGGTSAPIAPSSGSPRR104 pKa = 11.84SIWAKK109 pKa = 8.88LWSWRR114 pKa = 11.84LGYY117 pKa = 7.62PTTTLSAEE125 pKa = 4.22MADD128 pKa = 3.63ALAVGATTEE137 pKa = 4.11VEE139 pKa = 4.38EE140 pKa = 4.28EE141 pKa = 3.83ALAALDD147 pKa = 3.9EE148 pKa = 4.45QCNVHH153 pKa = 6.66YY154 pKa = 10.8GDD156 pKa = 4.22GQPPSFLGGDD166 pKa = 3.54HH167 pKa = 6.64NFTRR171 pKa = 11.84FAYY174 pKa = 10.17RR175 pKa = 11.84LSIKK179 pKa = 10.61AKK181 pKa = 10.5LKK183 pKa = 10.43FGYY186 pKa = 9.58DD187 pKa = 3.84LKK189 pKa = 10.43PTVANRR195 pKa = 11.84QVVHH199 pKa = 6.11EE200 pKa = 4.1WLVRR204 pKa = 11.84HH205 pKa = 5.45MRR207 pKa = 11.84DD208 pKa = 2.99MNVRR212 pKa = 11.84TTHH215 pKa = 6.68IARR218 pKa = 11.84VLPIAIPLVFVKK230 pKa = 10.55DD231 pKa = 3.93KK232 pKa = 11.05YY233 pKa = 11.21QLDD236 pKa = 3.91ADD238 pKa = 4.15NALAGAPMRR247 pKa = 11.84RR248 pKa = 11.84AAQEE252 pKa = 3.6SRR254 pKa = 11.84KK255 pKa = 7.84TVTEE259 pKa = 3.98RR260 pKa = 11.84WRR262 pKa = 11.84EE263 pKa = 3.75QRR265 pKa = 11.84TWWEE269 pKa = 4.22WLWGTSPRR277 pKa = 11.84PLTFTEE283 pKa = 4.22
MM1 pKa = 7.07IQVPSRR7 pKa = 11.84QGLQAIGGGSTEE19 pKa = 3.83QLRR22 pKa = 11.84SHH24 pKa = 7.04RR25 pKa = 11.84PTLLGLFEE33 pKa = 5.74DD34 pKa = 4.33GWMGYY39 pKa = 9.49PSALSPSTAGDD50 pKa = 3.57VEE52 pKa = 4.66VPKK55 pKa = 10.83AQAEE59 pKa = 4.6VPSNNARR66 pKa = 11.84GPLFGLGEE74 pKa = 4.43SPTAKK79 pKa = 10.0TPGEE83 pKa = 4.35TPFSQVGGTSAPIAPSSGSPRR104 pKa = 11.84SIWAKK109 pKa = 8.88LWSWRR114 pKa = 11.84LGYY117 pKa = 7.62PTTTLSAEE125 pKa = 4.22MADD128 pKa = 3.63ALAVGATTEE137 pKa = 4.11VEE139 pKa = 4.38EE140 pKa = 4.28EE141 pKa = 3.83ALAALDD147 pKa = 3.9EE148 pKa = 4.45QCNVHH153 pKa = 6.66YY154 pKa = 10.8GDD156 pKa = 4.22GQPPSFLGGDD166 pKa = 3.54HH167 pKa = 6.64NFTRR171 pKa = 11.84FAYY174 pKa = 10.17RR175 pKa = 11.84LSIKK179 pKa = 10.61AKK181 pKa = 10.5LKK183 pKa = 10.43FGYY186 pKa = 9.58DD187 pKa = 3.84LKK189 pKa = 10.43PTVANRR195 pKa = 11.84QVVHH199 pKa = 6.11EE200 pKa = 4.1WLVRR204 pKa = 11.84HH205 pKa = 5.45MRR207 pKa = 11.84DD208 pKa = 2.99MNVRR212 pKa = 11.84TTHH215 pKa = 6.68IARR218 pKa = 11.84VLPIAIPLVFVKK230 pKa = 10.55DD231 pKa = 3.93KK232 pKa = 11.05YY233 pKa = 11.21QLDD236 pKa = 3.91ADD238 pKa = 4.15NALAGAPMRR247 pKa = 11.84RR248 pKa = 11.84AAQEE252 pKa = 3.6SRR254 pKa = 11.84KK255 pKa = 7.84TVTEE259 pKa = 3.98RR260 pKa = 11.84WRR262 pKa = 11.84EE263 pKa = 3.75QRR265 pKa = 11.84TWWEE269 pKa = 4.22WLWGTSPRR277 pKa = 11.84PLTFTEE283 pKa = 4.22
Molecular weight: 31.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFV6|A0A1L3KFV6_9VIRU Tombus_P33 domain-containing protein OS=Changjiang tombus-like virus 4 OX=1922818 PE=4 SV=1
MM1 pKa = 7.62AKK3 pKa = 10.34GNRR6 pKa = 11.84SRR8 pKa = 11.84SKK10 pKa = 10.8RR11 pKa = 11.84KK12 pKa = 9.81GKK14 pKa = 10.4APNAGNTTRR23 pKa = 11.84AKK25 pKa = 10.2PALNLVRR32 pKa = 11.84RR33 pKa = 11.84SRR35 pKa = 11.84LLSLTPAEE43 pKa = 4.03RR44 pKa = 11.84KK45 pKa = 9.14YY46 pKa = 11.27AHH48 pKa = 6.9LVADD52 pKa = 4.42PCNGPLSAGIFGDD65 pKa = 3.78GSGGVISRR73 pKa = 11.84FEE75 pKa = 3.94TDD77 pKa = 4.21GIMGDD82 pKa = 3.79VAPTTASALVFVPAAAAGWTSGKK105 pKa = 9.74PSDD108 pKa = 4.45GDD110 pKa = 3.63APVWNGVTSSLIPGRR125 pKa = 11.84DD126 pKa = 3.61FLNANAGQFRR136 pKa = 11.84CLAACLRR143 pKa = 11.84IYY145 pKa = 10.22WPGTEE150 pKa = 4.88LNRR153 pKa = 11.84QGIVSNLQTTADD165 pKa = 3.14IVNNSNASVGAIRR178 pKa = 11.84ASSTYY183 pKa = 7.19VQRR186 pKa = 11.84MPEE189 pKa = 4.59DD190 pKa = 3.5YY191 pKa = 10.82TEE193 pKa = 4.95LKK195 pKa = 9.18WLPSEE200 pKa = 4.76YY201 pKa = 9.16EE202 pKa = 3.99LEE204 pKa = 3.98MRR206 pKa = 11.84APGVIAASQEE216 pKa = 3.84FSRR219 pKa = 11.84FSALVTTTSGVPSATPIRR237 pKa = 11.84WRR239 pKa = 11.84MVAVYY244 pKa = 8.94EE245 pKa = 3.98WVPRR249 pKa = 11.84VSTGLSSMNTAASVPAGSFQRR270 pKa = 11.84VTQALTSIGNWAYY283 pKa = 10.94VSSHH287 pKa = 5.08QAATAISSLLAGAHH301 pKa = 6.2AAGTLASGVATLTLGG316 pKa = 3.54
MM1 pKa = 7.62AKK3 pKa = 10.34GNRR6 pKa = 11.84SRR8 pKa = 11.84SKK10 pKa = 10.8RR11 pKa = 11.84KK12 pKa = 9.81GKK14 pKa = 10.4APNAGNTTRR23 pKa = 11.84AKK25 pKa = 10.2PALNLVRR32 pKa = 11.84RR33 pKa = 11.84SRR35 pKa = 11.84LLSLTPAEE43 pKa = 4.03RR44 pKa = 11.84KK45 pKa = 9.14YY46 pKa = 11.27AHH48 pKa = 6.9LVADD52 pKa = 4.42PCNGPLSAGIFGDD65 pKa = 3.78GSGGVISRR73 pKa = 11.84FEE75 pKa = 3.94TDD77 pKa = 4.21GIMGDD82 pKa = 3.79VAPTTASALVFVPAAAAGWTSGKK105 pKa = 9.74PSDD108 pKa = 4.45GDD110 pKa = 3.63APVWNGVTSSLIPGRR125 pKa = 11.84DD126 pKa = 3.61FLNANAGQFRR136 pKa = 11.84CLAACLRR143 pKa = 11.84IYY145 pKa = 10.22WPGTEE150 pKa = 4.88LNRR153 pKa = 11.84QGIVSNLQTTADD165 pKa = 3.14IVNNSNASVGAIRR178 pKa = 11.84ASSTYY183 pKa = 7.19VQRR186 pKa = 11.84MPEE189 pKa = 4.59DD190 pKa = 3.5YY191 pKa = 10.82TEE193 pKa = 4.95LKK195 pKa = 9.18WLPSEE200 pKa = 4.76YY201 pKa = 9.16EE202 pKa = 3.99LEE204 pKa = 3.98MRR206 pKa = 11.84APGVIAASQEE216 pKa = 3.84FSRR219 pKa = 11.84FSALVTTTSGVPSATPIRR237 pKa = 11.84WRR239 pKa = 11.84MVAVYY244 pKa = 8.94EE245 pKa = 3.98WVPRR249 pKa = 11.84VSTGLSSMNTAASVPAGSFQRR270 pKa = 11.84VTQALTSIGNWAYY283 pKa = 10.94VSSHH287 pKa = 5.08QAATAISSLLAGAHH301 pKa = 6.2AAGTLASGVATLTLGG316 pKa = 3.54
Molecular weight: 33.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1075 |
283 |
476 |
358.3 |
39.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.07 ± 1.262 | 1.209 ± 0.368 |
3.535 ± 0.27 | 4.744 ± 0.702 |
3.535 ± 0.49 | 8.837 ± 0.153 |
2.233 ± 0.562 | 3.814 ± 0.362 |
3.349 ± 0.327 | 8.0 ± 0.291 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.512 ± 0.341 | 3.721 ± 0.494 |
5.767 ± 0.718 | 3.535 ± 0.404 |
8.0 ± 0.772 | 7.535 ± 1.313 |
6.233 ± 1.062 | 7.163 ± 0.404 |
2.512 ± 0.343 | 2.698 ± 0.345 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
