
Canis familiaris papillomavirus 7
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Taupapillomavirus; Taupapillomavirus 1
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
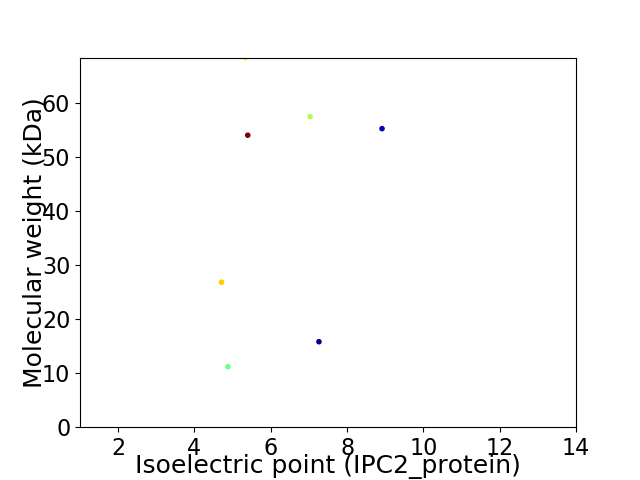
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
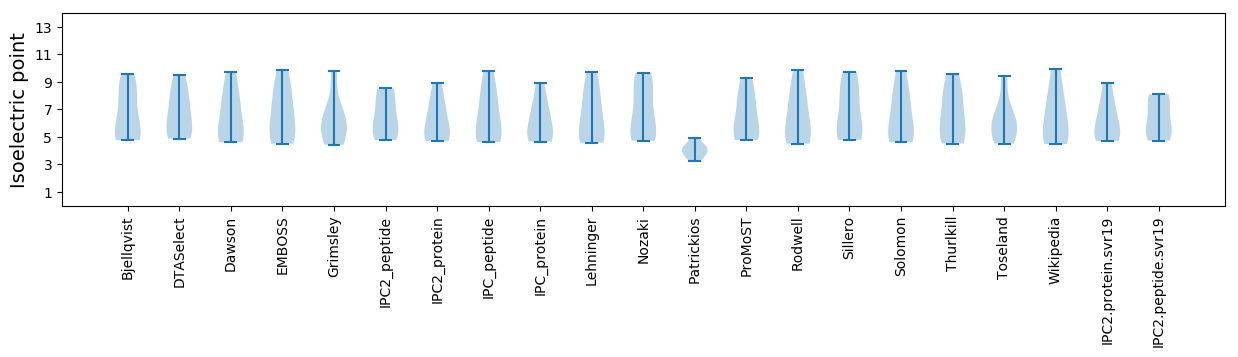
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C8YJK0|C8YJK0_9PAPI Replication protein E1 OS=Canis familiaris papillomavirus 7 OX=2759772 GN=E1 PE=3 SV=1
MM1 pKa = 7.75RR2 pKa = 11.84GSEE5 pKa = 4.07SDD7 pKa = 3.33VRR9 pKa = 11.84DD10 pKa = 3.33IVLDD14 pKa = 3.4LHH16 pKa = 6.79EE17 pKa = 5.51LVLPQNLLSSEE28 pKa = 4.11VLEE31 pKa = 4.54TEE33 pKa = 4.15EE34 pKa = 4.82EE35 pKa = 4.31EE36 pKa = 4.57PEE38 pKa = 4.32PEE40 pKa = 3.92PGRR43 pKa = 11.84YY44 pKa = 8.06RR45 pKa = 11.84VVTCCNLCHH54 pKa = 6.64SPLRR58 pKa = 11.84LFIEE62 pKa = 4.68SADD65 pKa = 3.71EE66 pKa = 4.14GQVRR70 pKa = 11.84IFHH73 pKa = 6.04QLLLDD78 pKa = 3.91GLGILCAVCYY88 pKa = 7.75RR89 pKa = 11.84THH91 pKa = 6.59CCDD94 pKa = 2.63GRR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 3.61
MM1 pKa = 7.75RR2 pKa = 11.84GSEE5 pKa = 4.07SDD7 pKa = 3.33VRR9 pKa = 11.84DD10 pKa = 3.33IVLDD14 pKa = 3.4LHH16 pKa = 6.79EE17 pKa = 5.51LVLPQNLLSSEE28 pKa = 4.11VLEE31 pKa = 4.54TEE33 pKa = 4.15EE34 pKa = 4.82EE35 pKa = 4.31EE36 pKa = 4.57PEE38 pKa = 4.32PEE40 pKa = 3.92PGRR43 pKa = 11.84YY44 pKa = 8.06RR45 pKa = 11.84VVTCCNLCHH54 pKa = 6.64SPLRR58 pKa = 11.84LFIEE62 pKa = 4.68SADD65 pKa = 3.71EE66 pKa = 4.14GQVRR70 pKa = 11.84IFHH73 pKa = 6.04QLLLDD78 pKa = 3.91GLGILCAVCYY88 pKa = 7.75RR89 pKa = 11.84THH91 pKa = 6.59CCDD94 pKa = 2.63GRR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 3.61
Molecular weight: 11.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C8YJK2|C8YJK2_9PAPI E4 (Fragment) OS=Canis familiaris papillomavirus 7 OX=2759772 PE=4 SV=1
MM1 pKa = 7.22MEE3 pKa = 4.0SLGRR7 pKa = 11.84SFDD10 pKa = 3.58VLQQVMLVHH19 pKa = 5.99YY20 pKa = 9.52EE21 pKa = 3.98RR22 pKa = 11.84GSKK25 pKa = 10.15RR26 pKa = 11.84LSDD29 pKa = 4.62QIIFWEE35 pKa = 4.16AVRR38 pKa = 11.84RR39 pKa = 11.84EE40 pKa = 3.98NALLHH45 pKa = 5.37YY46 pKa = 10.26ARR48 pKa = 11.84KK49 pKa = 9.36QRR51 pKa = 11.84IARR54 pKa = 11.84LGYY57 pKa = 8.53TPVPSSATSEE67 pKa = 3.97AAAKK71 pKa = 10.16NAIKK75 pKa = 8.25MTLLLTSLNEE85 pKa = 3.98SEE87 pKa = 5.2YY88 pKa = 11.04KK89 pKa = 10.62DD90 pKa = 4.47EE91 pKa = 4.57PWTMSDD97 pKa = 3.17TSLEE101 pKa = 3.97MLQCKK106 pKa = 9.84PKK108 pKa = 10.74GCFKK112 pKa = 10.83KK113 pKa = 10.48RR114 pKa = 11.84GFTVHH119 pKa = 6.23VLFDD123 pKa = 4.31GDD125 pKa = 4.04PGNSFPYY132 pKa = 9.46TSWGEE137 pKa = 3.67IYY139 pKa = 10.55YY140 pKa = 10.79QNLNDD145 pKa = 3.63KK146 pKa = 9.25WIKK149 pKa = 9.84TEE151 pKa = 4.06GKK153 pKa = 9.59VDD155 pKa = 3.76YY156 pKa = 10.92DD157 pKa = 3.27GLYY160 pKa = 8.86YY161 pKa = 10.35TDD163 pKa = 4.23EE164 pKa = 5.85DD165 pKa = 4.5GEE167 pKa = 4.18KK168 pKa = 10.44LYY170 pKa = 11.51YY171 pKa = 9.31EE172 pKa = 4.7TFAKK176 pKa = 10.62DD177 pKa = 3.11AMRR180 pKa = 11.84FGTTGMWKK188 pKa = 9.11VCYY191 pKa = 9.97KK192 pKa = 9.97STVLSALVSSSGTPAPSSVLWSQEE216 pKa = 3.17WARR219 pKa = 11.84EE220 pKa = 4.06GGLEE224 pKa = 4.14TQNPPVPATSDD235 pKa = 3.05SRR237 pKa = 11.84QPEE240 pKa = 4.27SSVSHH245 pKa = 6.8ASGTSTGSFARR256 pKa = 11.84PGGEE260 pKa = 3.98ACSSTPYY267 pKa = 10.27RR268 pKa = 11.84RR269 pKa = 11.84GQEE272 pKa = 3.84GGRR275 pKa = 11.84GRR277 pKa = 11.84GRR279 pKa = 11.84GRR281 pKa = 11.84GRR283 pKa = 11.84GRR285 pKa = 11.84GRR287 pKa = 11.84GRR289 pKa = 11.84PAGRR293 pKa = 11.84QEE295 pKa = 4.18GTPGSPSRR303 pKa = 11.84STLPCSVLGRR313 pKa = 11.84QGAQKK318 pKa = 9.94RR319 pKa = 11.84QRR321 pKa = 11.84EE322 pKa = 4.04ASPNRR327 pKa = 11.84GSEE330 pKa = 3.74QDD332 pKa = 3.06RR333 pKa = 11.84GGGEE337 pKa = 4.19GGQGEE342 pKa = 4.73SGEE345 pKa = 4.44SASNEE350 pKa = 4.0PEE352 pKa = 4.17STNSPRR358 pKa = 11.84LPRR361 pKa = 11.84LSTPGSVSRR370 pKa = 11.84VGGPCRR376 pKa = 11.84PSPRR380 pKa = 11.84KK381 pKa = 9.31RR382 pKa = 11.84APPRR386 pKa = 11.84LSPAVSYY393 pKa = 10.38RR394 pKa = 11.84QVGGGCGGSEE404 pKa = 4.2GQGKK408 pKa = 9.16LLGGCTEE415 pKa = 4.49GEE417 pKa = 4.17DD418 pKa = 3.88RR419 pKa = 11.84PTPILIFRR427 pKa = 11.84GDD429 pKa = 3.76GNTQKK434 pKa = 10.32CWRR437 pKa = 11.84RR438 pKa = 11.84RR439 pKa = 11.84LRR441 pKa = 11.84IRR443 pKa = 11.84HH444 pKa = 5.33SSRR447 pKa = 11.84YY448 pKa = 7.85MCASTGFSWTIPRR461 pKa = 11.84GPNTVGRR468 pKa = 11.84HH469 pKa = 4.45RR470 pKa = 11.84MLLAFNSDD478 pKa = 3.26SQRR481 pKa = 11.84AAFLEE486 pKa = 4.6TVTIPRR492 pKa = 11.84GLEE495 pKa = 3.69FSEE498 pKa = 5.88GYY500 pKa = 10.28LDD502 pKa = 5.12SII504 pKa = 5.19
MM1 pKa = 7.22MEE3 pKa = 4.0SLGRR7 pKa = 11.84SFDD10 pKa = 3.58VLQQVMLVHH19 pKa = 5.99YY20 pKa = 9.52EE21 pKa = 3.98RR22 pKa = 11.84GSKK25 pKa = 10.15RR26 pKa = 11.84LSDD29 pKa = 4.62QIIFWEE35 pKa = 4.16AVRR38 pKa = 11.84RR39 pKa = 11.84EE40 pKa = 3.98NALLHH45 pKa = 5.37YY46 pKa = 10.26ARR48 pKa = 11.84KK49 pKa = 9.36QRR51 pKa = 11.84IARR54 pKa = 11.84LGYY57 pKa = 8.53TPVPSSATSEE67 pKa = 3.97AAAKK71 pKa = 10.16NAIKK75 pKa = 8.25MTLLLTSLNEE85 pKa = 3.98SEE87 pKa = 5.2YY88 pKa = 11.04KK89 pKa = 10.62DD90 pKa = 4.47EE91 pKa = 4.57PWTMSDD97 pKa = 3.17TSLEE101 pKa = 3.97MLQCKK106 pKa = 9.84PKK108 pKa = 10.74GCFKK112 pKa = 10.83KK113 pKa = 10.48RR114 pKa = 11.84GFTVHH119 pKa = 6.23VLFDD123 pKa = 4.31GDD125 pKa = 4.04PGNSFPYY132 pKa = 9.46TSWGEE137 pKa = 3.67IYY139 pKa = 10.55YY140 pKa = 10.79QNLNDD145 pKa = 3.63KK146 pKa = 9.25WIKK149 pKa = 9.84TEE151 pKa = 4.06GKK153 pKa = 9.59VDD155 pKa = 3.76YY156 pKa = 10.92DD157 pKa = 3.27GLYY160 pKa = 8.86YY161 pKa = 10.35TDD163 pKa = 4.23EE164 pKa = 5.85DD165 pKa = 4.5GEE167 pKa = 4.18KK168 pKa = 10.44LYY170 pKa = 11.51YY171 pKa = 9.31EE172 pKa = 4.7TFAKK176 pKa = 10.62DD177 pKa = 3.11AMRR180 pKa = 11.84FGTTGMWKK188 pKa = 9.11VCYY191 pKa = 9.97KK192 pKa = 9.97STVLSALVSSSGTPAPSSVLWSQEE216 pKa = 3.17WARR219 pKa = 11.84EE220 pKa = 4.06GGLEE224 pKa = 4.14TQNPPVPATSDD235 pKa = 3.05SRR237 pKa = 11.84QPEE240 pKa = 4.27SSVSHH245 pKa = 6.8ASGTSTGSFARR256 pKa = 11.84PGGEE260 pKa = 3.98ACSSTPYY267 pKa = 10.27RR268 pKa = 11.84RR269 pKa = 11.84GQEE272 pKa = 3.84GGRR275 pKa = 11.84GRR277 pKa = 11.84GRR279 pKa = 11.84GRR281 pKa = 11.84GRR283 pKa = 11.84GRR285 pKa = 11.84GRR287 pKa = 11.84GRR289 pKa = 11.84PAGRR293 pKa = 11.84QEE295 pKa = 4.18GTPGSPSRR303 pKa = 11.84STLPCSVLGRR313 pKa = 11.84QGAQKK318 pKa = 9.94RR319 pKa = 11.84QRR321 pKa = 11.84EE322 pKa = 4.04ASPNRR327 pKa = 11.84GSEE330 pKa = 3.74QDD332 pKa = 3.06RR333 pKa = 11.84GGGEE337 pKa = 4.19GGQGEE342 pKa = 4.73SGEE345 pKa = 4.44SASNEE350 pKa = 4.0PEE352 pKa = 4.17STNSPRR358 pKa = 11.84LPRR361 pKa = 11.84LSTPGSVSRR370 pKa = 11.84VGGPCRR376 pKa = 11.84PSPRR380 pKa = 11.84KK381 pKa = 9.31RR382 pKa = 11.84APPRR386 pKa = 11.84LSPAVSYY393 pKa = 10.38RR394 pKa = 11.84QVGGGCGGSEE404 pKa = 4.2GQGKK408 pKa = 9.16LLGGCTEE415 pKa = 4.49GEE417 pKa = 4.17DD418 pKa = 3.88RR419 pKa = 11.84PTPILIFRR427 pKa = 11.84GDD429 pKa = 3.76GNTQKK434 pKa = 10.32CWRR437 pKa = 11.84RR438 pKa = 11.84RR439 pKa = 11.84LRR441 pKa = 11.84IRR443 pKa = 11.84HH444 pKa = 5.33SSRR447 pKa = 11.84YY448 pKa = 7.85MCASTGFSWTIPRR461 pKa = 11.84GPNTVGRR468 pKa = 11.84HH469 pKa = 4.45RR470 pKa = 11.84MLLAFNSDD478 pKa = 3.26SQRR481 pKa = 11.84AAFLEE486 pKa = 4.6TVTIPRR492 pKa = 11.84GLEE495 pKa = 3.69FSEE498 pKa = 5.88GYY500 pKa = 10.28LDD502 pKa = 5.12SII504 pKa = 5.19
Molecular weight: 55.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2603 |
98 |
605 |
371.9 |
41.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.34 ± 0.324 | 2.536 ± 0.737 |
5.839 ± 0.708 | 6.531 ± 0.632 |
4.034 ± 0.684 | 8.529 ± 1.164 |
2.113 ± 0.356 | 3.765 ± 0.732 |
4.495 ± 0.701 | 8.759 ± 1.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.614 ± 0.396 | 3.803 ± 0.744 |
6.954 ± 1.039 | 4.495 ± 0.561 |
6.723 ± 1.055 | 8.951 ± 0.903 |
5.839 ± 0.677 | 5.532 ± 0.717 |
1.268 ± 0.329 | 2.881 ± 0.415 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
