
Beijerinckiaceae bacterium RH AL1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Beijerinckiaceae; unclassified Beijerinckiaceae
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
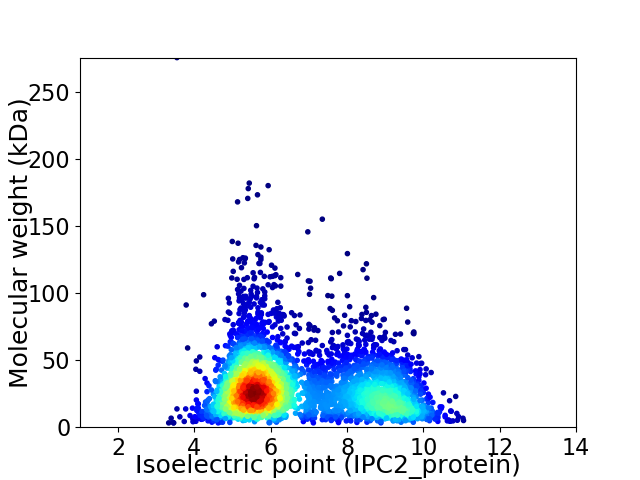
Virtual 2D-PAGE plot for 4238 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5E4NUA7|A0A5E4NUA7_9RHIZ RNA polymerase-binding transcription factor DksA OS=Beijerinckiaceae bacterium RH AL1 OX=2572036 GN=dksA PE=3 SV=1
MM1 pKa = 7.49SLFNSMRR8 pKa = 11.84TAVSGMAAQSNALSAISDD26 pKa = 4.06NIANSSTVGYY36 pKa = 10.44KK37 pKa = 10.06DD38 pKa = 3.39AQAQFEE44 pKa = 4.58TVLDD48 pKa = 3.82QNATSEE54 pKa = 4.39YY55 pKa = 10.2EE56 pKa = 4.15SGGVQTDD63 pKa = 2.53IRR65 pKa = 11.84YY66 pKa = 8.93GVSTQGTLEE75 pKa = 4.28TTTSSTDD82 pKa = 3.11MAINGNGFFVVSNGGQGTYY101 pKa = 8.06LTRR104 pKa = 11.84AGSFVPDD111 pKa = 3.32STGNLVNTAGYY122 pKa = 9.53QLQGYY127 pKa = 8.87KK128 pKa = 10.03INPDD132 pKa = 3.02GTTSSTLSTVNINNQTLQAAASTTGTLTANLPSTATAVAANTPGTSNSTSTTYY185 pKa = 10.32TDD187 pKa = 3.14KK188 pKa = 11.37TSITVYY194 pKa = 11.02DD195 pKa = 4.22DD196 pKa = 4.79LGTSHH201 pKa = 7.33VMDD204 pKa = 4.14VYY206 pKa = 10.26LTKK209 pKa = 10.41TGNNTWEE216 pKa = 3.91AAAYY220 pKa = 6.94PQADD224 pKa = 3.47ASASGGFPYY233 pKa = 10.6SSGGTTDD240 pKa = 4.77AVATPTTLKK249 pKa = 10.53FDD251 pKa = 3.73PTTGNMLNTPVTMNIQIPGGQAGQTVAVNLSATTQLASSFAVTAGTADD299 pKa = 3.33GNAPSKK305 pKa = 10.71LSNVKK310 pKa = 9.94IGTDD314 pKa = 3.5GTVTEE319 pKa = 5.5VYY321 pKa = 10.82ASGYY325 pKa = 9.59QLAAYY330 pKa = 7.7KK331 pKa = 10.33IPLATVEE338 pKa = 4.58SPTNLTNLSGNVYY351 pKa = 10.0QVSQNSGPMVLSTATTNGTGSILADD376 pKa = 3.51TLEE379 pKa = 4.35EE380 pKa = 4.33STVDD384 pKa = 4.44LATEE388 pKa = 4.53LTNMISAQRR397 pKa = 11.84SYY399 pKa = 10.86EE400 pKa = 4.04ANSKK404 pKa = 9.58VLQAASDD411 pKa = 3.9LLGDD415 pKa = 4.53LNRR418 pKa = 11.84LTTNN422 pKa = 3.32
MM1 pKa = 7.49SLFNSMRR8 pKa = 11.84TAVSGMAAQSNALSAISDD26 pKa = 4.06NIANSSTVGYY36 pKa = 10.44KK37 pKa = 10.06DD38 pKa = 3.39AQAQFEE44 pKa = 4.58TVLDD48 pKa = 3.82QNATSEE54 pKa = 4.39YY55 pKa = 10.2EE56 pKa = 4.15SGGVQTDD63 pKa = 2.53IRR65 pKa = 11.84YY66 pKa = 8.93GVSTQGTLEE75 pKa = 4.28TTTSSTDD82 pKa = 3.11MAINGNGFFVVSNGGQGTYY101 pKa = 8.06LTRR104 pKa = 11.84AGSFVPDD111 pKa = 3.32STGNLVNTAGYY122 pKa = 9.53QLQGYY127 pKa = 8.87KK128 pKa = 10.03INPDD132 pKa = 3.02GTTSSTLSTVNINNQTLQAAASTTGTLTANLPSTATAVAANTPGTSNSTSTTYY185 pKa = 10.32TDD187 pKa = 3.14KK188 pKa = 11.37TSITVYY194 pKa = 11.02DD195 pKa = 4.22DD196 pKa = 4.79LGTSHH201 pKa = 7.33VMDD204 pKa = 4.14VYY206 pKa = 10.26LTKK209 pKa = 10.41TGNNTWEE216 pKa = 3.91AAAYY220 pKa = 6.94PQADD224 pKa = 3.47ASASGGFPYY233 pKa = 10.6SSGGTTDD240 pKa = 4.77AVATPTTLKK249 pKa = 10.53FDD251 pKa = 3.73PTTGNMLNTPVTMNIQIPGGQAGQTVAVNLSATTQLASSFAVTAGTADD299 pKa = 3.33GNAPSKK305 pKa = 10.71LSNVKK310 pKa = 9.94IGTDD314 pKa = 3.5GTVTEE319 pKa = 5.5VYY321 pKa = 10.82ASGYY325 pKa = 9.59QLAAYY330 pKa = 7.7KK331 pKa = 10.33IPLATVEE338 pKa = 4.58SPTNLTNLSGNVYY351 pKa = 10.0QVSQNSGPMVLSTATTNGTGSILADD376 pKa = 3.51TLEE379 pKa = 4.35EE380 pKa = 4.33STVDD384 pKa = 4.44LATEE388 pKa = 4.53LTNMISAQRR397 pKa = 11.84SYY399 pKa = 10.86EE400 pKa = 4.04ANSKK404 pKa = 9.58VLQAASDD411 pKa = 3.9LLGDD415 pKa = 4.53LNRR418 pKa = 11.84LTTNN422 pKa = 3.32
Molecular weight: 43.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5E4NW26|A0A5E4NW26_9RHIZ Dihydrofolate synthase/folylpolyglutamate synthase OS=Beijerinckiaceae bacterium RH AL1 OX=2572036 GN=RHAL1_01252 PE=3 SV=1
MM1 pKa = 7.67LKK3 pKa = 9.02TAIAASIVLGSVSFASAMPMASSVQTGASLPVVKK37 pKa = 10.34AAVIVKK43 pKa = 9.72KK44 pKa = 9.13VVRR47 pKa = 11.84RR48 pKa = 11.84PPVRR52 pKa = 11.84RR53 pKa = 11.84TVVRR57 pKa = 11.84KK58 pKa = 8.19TIIRR62 pKa = 11.84RR63 pKa = 3.52
MM1 pKa = 7.67LKK3 pKa = 9.02TAIAASIVLGSVSFASAMPMASSVQTGASLPVVKK37 pKa = 10.34AAVIVKK43 pKa = 9.72KK44 pKa = 9.13VVRR47 pKa = 11.84RR48 pKa = 11.84PPVRR52 pKa = 11.84RR53 pKa = 11.84TVVRR57 pKa = 11.84KK58 pKa = 8.19TIIRR62 pKa = 11.84RR63 pKa = 3.52
Molecular weight: 6.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1273430 |
29 |
2807 |
300.5 |
32.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.971 ± 0.061 | 0.892 ± 0.013 |
5.82 ± 0.032 | 5.278 ± 0.035 |
3.618 ± 0.028 | 8.487 ± 0.039 |
2.13 ± 0.017 | 4.761 ± 0.027 |
3.237 ± 0.035 | 10.013 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.288 ± 0.017 | 2.201 ± 0.025 |
5.663 ± 0.034 | 2.813 ± 0.022 |
7.37 ± 0.047 | 5.093 ± 0.029 |
5.419 ± 0.038 | 7.548 ± 0.031 |
1.24 ± 0.015 | 2.158 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
