
Marinicella litoralis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Alcanivoracaceae; Marinicella
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
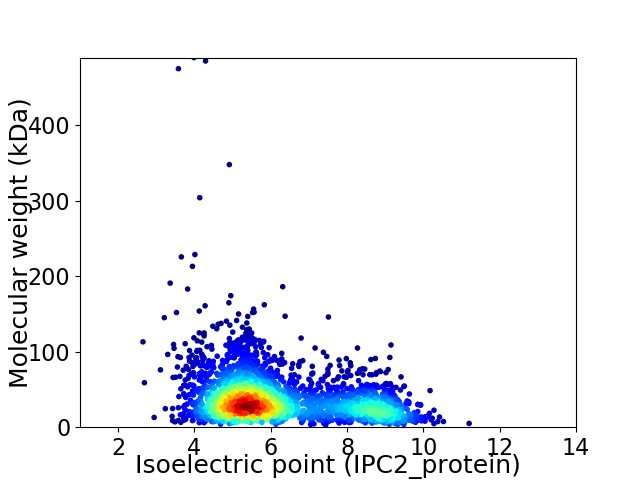
Virtual 2D-PAGE plot for 2897 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R6XMW7|A0A4R6XMW7_9GAMM Putative ABC transport system permease protein OS=Marinicella litoralis OX=644220 GN=C8D91_2016 PE=4 SV=1
MM1 pKa = 7.55FFLCFILLSLISLNVTAQNYY21 pKa = 9.3FSDD24 pKa = 3.81VSNQAGVAINHH35 pKa = 5.32QLKK38 pKa = 10.63SGEE41 pKa = 4.18LGMGQGWVDD50 pKa = 3.64VNNDD54 pKa = 3.15GYY56 pKa = 11.37EE57 pKa = 4.12DD58 pKa = 4.92LMLTNQSGEE67 pKa = 3.83NYY69 pKa = 10.51LFINQQDD76 pKa = 3.55GSFVEE81 pKa = 4.23MAQYY85 pKa = 11.47NDD87 pKa = 2.83IKK89 pKa = 11.13LPLSQDD95 pKa = 3.26FGVSVTDD102 pKa = 3.78FNNDD106 pKa = 2.48GFQDD110 pKa = 3.81LFIASWGVNKK120 pKa = 10.21LLKK123 pKa = 10.39NINGSSFLDD132 pKa = 3.66VSDD135 pKa = 3.94EE136 pKa = 4.03MGFVDD141 pKa = 4.36NSYY144 pKa = 11.07SIASTWADD152 pKa = 2.94INQDD156 pKa = 3.04GWLDD160 pKa = 3.92LYY162 pKa = 10.74VVNYY166 pKa = 10.36SVEE169 pKa = 4.08NDD171 pKa = 2.98SDD173 pKa = 3.76EE174 pKa = 4.71ADD176 pKa = 3.41RR177 pKa = 11.84MYY179 pKa = 11.32LNNQGANFIDD189 pKa = 4.05VSSDD193 pKa = 3.03LTPFTNLTKK202 pKa = 10.6HH203 pKa = 5.5GLAVQFIDD211 pKa = 4.73FDD213 pKa = 3.86MDD215 pKa = 3.47NDD217 pKa = 3.23MDD219 pKa = 4.9LYY221 pKa = 11.14VVNDD225 pKa = 3.7KK226 pKa = 11.4LEE228 pKa = 4.02QNTLWRR234 pKa = 11.84NDD236 pKa = 3.45GPGSSACGVYY246 pKa = 10.34VCFTDD251 pKa = 5.29VSALTQTNRR260 pKa = 11.84AVFGMGIAIADD271 pKa = 3.74YY272 pKa = 11.23DD273 pKa = 4.43LDD275 pKa = 4.97GDD277 pKa = 3.83FDD279 pKa = 5.77FYY281 pKa = 11.08FSSIAEE287 pKa = 4.05QVLLQSQISQGSAVFVEE304 pKa = 4.45QSNPAGLNFNATGWATIFADD324 pKa = 4.43FNNDD328 pKa = 3.45QFPDD332 pKa = 3.7AYY334 pKa = 10.94LATANNTPSKK344 pKa = 10.49RR345 pKa = 11.84DD346 pKa = 3.27RR347 pKa = 11.84LYY349 pKa = 11.45LNNQSQQFTDD359 pKa = 3.52VTALSGIEE367 pKa = 4.09DD368 pKa = 4.52LYY370 pKa = 9.13MTVGAAKK377 pKa = 10.54GDD379 pKa = 3.63FDD381 pKa = 5.55NDD383 pKa = 3.57GKK385 pKa = 11.42VDD387 pKa = 4.03LVVGNWGEE395 pKa = 3.96NYY397 pKa = 9.32MLYY400 pKa = 10.87KK401 pKa = 9.84NTSLDD406 pKa = 3.36ANNWIKK412 pKa = 10.61IKK414 pKa = 10.82LIGGSGINQLAIGSKK429 pKa = 9.81VAVNTSNGQEE439 pKa = 4.17LVDD442 pKa = 3.65QLVSGGSHH450 pKa = 6.28GAGNSLVLHH459 pKa = 6.63FGLADD464 pKa = 3.53EE465 pKa = 5.29TIEE468 pKa = 4.29SVKK471 pKa = 9.45ITWPNGIISLLSGLSANQMHH491 pKa = 6.97TLTYY495 pKa = 9.57PGDD498 pKa = 4.24GIFMQGFEE506 pKa = 4.14
MM1 pKa = 7.55FFLCFILLSLISLNVTAQNYY21 pKa = 9.3FSDD24 pKa = 3.81VSNQAGVAINHH35 pKa = 5.32QLKK38 pKa = 10.63SGEE41 pKa = 4.18LGMGQGWVDD50 pKa = 3.64VNNDD54 pKa = 3.15GYY56 pKa = 11.37EE57 pKa = 4.12DD58 pKa = 4.92LMLTNQSGEE67 pKa = 3.83NYY69 pKa = 10.51LFINQQDD76 pKa = 3.55GSFVEE81 pKa = 4.23MAQYY85 pKa = 11.47NDD87 pKa = 2.83IKK89 pKa = 11.13LPLSQDD95 pKa = 3.26FGVSVTDD102 pKa = 3.78FNNDD106 pKa = 2.48GFQDD110 pKa = 3.81LFIASWGVNKK120 pKa = 10.21LLKK123 pKa = 10.39NINGSSFLDD132 pKa = 3.66VSDD135 pKa = 3.94EE136 pKa = 4.03MGFVDD141 pKa = 4.36NSYY144 pKa = 11.07SIASTWADD152 pKa = 2.94INQDD156 pKa = 3.04GWLDD160 pKa = 3.92LYY162 pKa = 10.74VVNYY166 pKa = 10.36SVEE169 pKa = 4.08NDD171 pKa = 2.98SDD173 pKa = 3.76EE174 pKa = 4.71ADD176 pKa = 3.41RR177 pKa = 11.84MYY179 pKa = 11.32LNNQGANFIDD189 pKa = 4.05VSSDD193 pKa = 3.03LTPFTNLTKK202 pKa = 10.6HH203 pKa = 5.5GLAVQFIDD211 pKa = 4.73FDD213 pKa = 3.86MDD215 pKa = 3.47NDD217 pKa = 3.23MDD219 pKa = 4.9LYY221 pKa = 11.14VVNDD225 pKa = 3.7KK226 pKa = 11.4LEE228 pKa = 4.02QNTLWRR234 pKa = 11.84NDD236 pKa = 3.45GPGSSACGVYY246 pKa = 10.34VCFTDD251 pKa = 5.29VSALTQTNRR260 pKa = 11.84AVFGMGIAIADD271 pKa = 3.74YY272 pKa = 11.23DD273 pKa = 4.43LDD275 pKa = 4.97GDD277 pKa = 3.83FDD279 pKa = 5.77FYY281 pKa = 11.08FSSIAEE287 pKa = 4.05QVLLQSQISQGSAVFVEE304 pKa = 4.45QSNPAGLNFNATGWATIFADD324 pKa = 4.43FNNDD328 pKa = 3.45QFPDD332 pKa = 3.7AYY334 pKa = 10.94LATANNTPSKK344 pKa = 10.49RR345 pKa = 11.84DD346 pKa = 3.27RR347 pKa = 11.84LYY349 pKa = 11.45LNNQSQQFTDD359 pKa = 3.52VTALSGIEE367 pKa = 4.09DD368 pKa = 4.52LYY370 pKa = 9.13MTVGAAKK377 pKa = 10.54GDD379 pKa = 3.63FDD381 pKa = 5.55NDD383 pKa = 3.57GKK385 pKa = 11.42VDD387 pKa = 4.03LVVGNWGEE395 pKa = 3.96NYY397 pKa = 9.32MLYY400 pKa = 10.87KK401 pKa = 9.84NTSLDD406 pKa = 3.36ANNWIKK412 pKa = 10.61IKK414 pKa = 10.82LIGGSGINQLAIGSKK429 pKa = 9.81VAVNTSNGQEE439 pKa = 4.17LVDD442 pKa = 3.65QLVSGGSHH450 pKa = 6.28GAGNSLVLHH459 pKa = 6.63FGLADD464 pKa = 3.53EE465 pKa = 5.29TIEE468 pKa = 4.29SVKK471 pKa = 9.45ITWPNGIISLLSGLSANQMHH491 pKa = 6.97TLTYY495 pKa = 9.57PGDD498 pKa = 4.24GIFMQGFEE506 pKa = 4.14
Molecular weight: 55.45 kDa
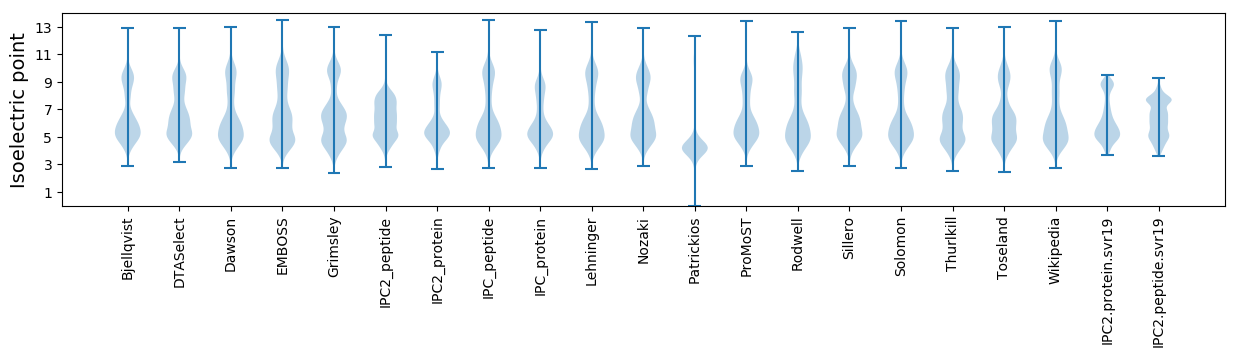
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R6XV76|A0A4R6XV76_9GAMM RND family efflux transporter MFP subunit OS=Marinicella litoralis OX=644220 GN=C8D91_0928 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 6.05GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.55TAGGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.8RR41 pKa = 11.84LAAA44 pKa = 4.42
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 6.05GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.55TAGGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.8RR41 pKa = 11.84LAAA44 pKa = 4.42
Molecular weight: 4.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1015597 |
26 |
4547 |
350.6 |
39.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.047 ± 0.046 | 0.94 ± 0.015 |
5.896 ± 0.041 | 5.589 ± 0.05 |
4.373 ± 0.037 | 6.738 ± 0.047 |
2.397 ± 0.026 | 6.722 ± 0.031 |
5.635 ± 0.056 | 9.972 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.707 ± 0.027 | 5.172 ± 0.044 |
3.847 ± 0.03 | 5.02 ± 0.038 |
3.689 ± 0.033 | 6.579 ± 0.044 |
5.433 ± 0.045 | 6.804 ± 0.043 |
1.325 ± 0.018 | 3.117 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
