
Circovirus-like genome RW-B
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
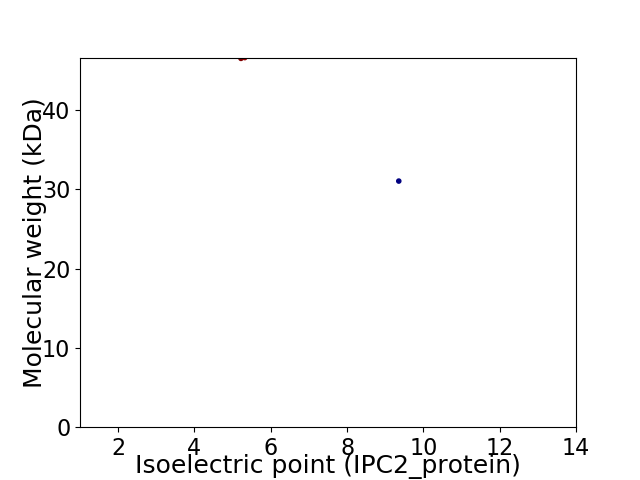
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
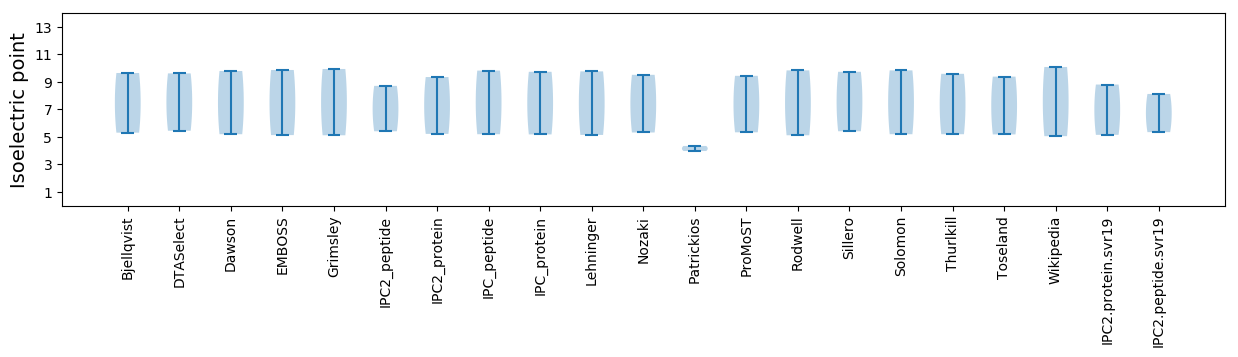
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C6GIH8|C6GIH8_9VIRU Coat protein OS=Circovirus-like genome RW-B OX=642252 PE=4 SV=1
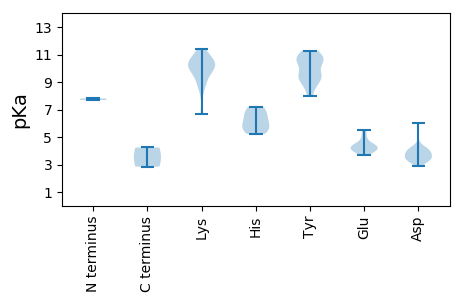
MM1 pKa = 7.81DD2 pKa = 4.28VDD4 pKa = 3.96TTPVSRR10 pKa = 11.84RR11 pKa = 11.84GRR13 pKa = 11.84PSSSSSSSSTGEE25 pKa = 3.66RR26 pKa = 11.84FTARR30 pKa = 11.84TFCVTVNNRR39 pKa = 11.84DD40 pKa = 3.42EE41 pKa = 5.54VEE43 pKa = 4.36QNWPNAVQEE52 pKa = 4.07PSEE55 pKa = 4.16EE56 pKa = 3.82FVEE59 pKa = 4.46RR60 pKa = 11.84VRR62 pKa = 11.84YY63 pKa = 8.49VVWQRR68 pKa = 11.84EE69 pKa = 4.24LAPEE73 pKa = 4.26TGRR76 pKa = 11.84VHH78 pKa = 5.23IQAYY82 pKa = 9.44VEE84 pKa = 4.51LYY86 pKa = 7.99MAAGIVMLKK95 pKa = 10.4RR96 pKa = 11.84LFNCPTMHH104 pKa = 5.61VEE106 pKa = 4.5KK107 pKa = 10.76RR108 pKa = 11.84RR109 pKa = 11.84GTQDD113 pKa = 2.9QARR116 pKa = 11.84NYY118 pKa = 9.97CMKK121 pKa = 10.21EE122 pKa = 3.95DD123 pKa = 3.97TRR125 pKa = 11.84DD126 pKa = 3.83GPDD129 pKa = 3.42GGPFEE134 pKa = 4.72YY135 pKa = 8.77GTYY138 pKa = 10.75SKK140 pKa = 9.96TPGNGQGKK148 pKa = 10.3RR149 pKa = 11.84NDD151 pKa = 3.3LTDD154 pKa = 3.44AVEE157 pKa = 4.0ALEE160 pKa = 4.42AGGVEE165 pKa = 4.33AVVSKK170 pKa = 11.01HH171 pKa = 5.41PNTYY175 pKa = 9.13VRR177 pKa = 11.84YY178 pKa = 9.29HH179 pKa = 6.61RR180 pKa = 11.84GIHH183 pKa = 6.31ALYY186 pKa = 9.48QAKK189 pKa = 10.52LEE191 pKa = 4.95AIASRR196 pKa = 11.84QQRR199 pKa = 11.84NVKK202 pKa = 8.46TAVFYY207 pKa = 11.11GEE209 pKa = 4.42PGTGKK214 pKa = 6.71THH216 pKa = 7.2CAFEE220 pKa = 4.16LARR223 pKa = 11.84ITGEE227 pKa = 3.84EE228 pKa = 4.22IYY230 pKa = 10.59ILNAPAGRR238 pKa = 11.84NQSVWFNGYY247 pKa = 8.23QNQKK251 pKa = 9.73ILVVDD256 pKa = 4.13EE257 pKa = 4.97MNGDD261 pKa = 3.97WIGWQLLLRR270 pKa = 11.84MTDD273 pKa = 4.21KK274 pKa = 11.41YY275 pKa = 11.03PLQCQTKK282 pKa = 10.07GGMVWAMWEE291 pKa = 4.04IVIFTSNAHH300 pKa = 5.64WEE302 pKa = 4.02DD303 pKa = 3.36WYY305 pKa = 10.38PYY307 pKa = 9.7YY308 pKa = 10.98AGGMDD313 pKa = 3.31KK314 pKa = 10.96AALKK318 pKa = 10.47RR319 pKa = 11.84RR320 pKa = 11.84IHH322 pKa = 6.1KK323 pKa = 9.25VVKK326 pKa = 10.17FYY328 pKa = 11.25GSEE331 pKa = 3.78ASGNFTKK338 pKa = 10.33KK339 pKa = 10.74YY340 pKa = 9.53YY341 pKa = 10.91DD342 pKa = 4.07PDD344 pKa = 3.64LPEE347 pKa = 5.21ANWTVATTDD356 pKa = 3.37VIATFLRR363 pKa = 11.84NGTVRR368 pKa = 11.84PHH370 pKa = 5.92TDD372 pKa = 2.99EE373 pKa = 4.26EE374 pKa = 4.71LEE376 pKa = 4.15EE377 pKa = 4.41VEE379 pKa = 4.33DD380 pKa = 4.2VVMNTQQEE388 pKa = 4.58SQGTQTTEE396 pKa = 3.79LVEE399 pKa = 5.35DD400 pKa = 4.42SDD402 pKa = 6.05DD403 pKa = 3.84EE404 pKa = 4.21MSEE407 pKa = 4.29TFF409 pKa = 4.24
MM1 pKa = 7.81DD2 pKa = 4.28VDD4 pKa = 3.96TTPVSRR10 pKa = 11.84RR11 pKa = 11.84GRR13 pKa = 11.84PSSSSSSSSTGEE25 pKa = 3.66RR26 pKa = 11.84FTARR30 pKa = 11.84TFCVTVNNRR39 pKa = 11.84DD40 pKa = 3.42EE41 pKa = 5.54VEE43 pKa = 4.36QNWPNAVQEE52 pKa = 4.07PSEE55 pKa = 4.16EE56 pKa = 3.82FVEE59 pKa = 4.46RR60 pKa = 11.84VRR62 pKa = 11.84YY63 pKa = 8.49VVWQRR68 pKa = 11.84EE69 pKa = 4.24LAPEE73 pKa = 4.26TGRR76 pKa = 11.84VHH78 pKa = 5.23IQAYY82 pKa = 9.44VEE84 pKa = 4.51LYY86 pKa = 7.99MAAGIVMLKK95 pKa = 10.4RR96 pKa = 11.84LFNCPTMHH104 pKa = 5.61VEE106 pKa = 4.5KK107 pKa = 10.76RR108 pKa = 11.84RR109 pKa = 11.84GTQDD113 pKa = 2.9QARR116 pKa = 11.84NYY118 pKa = 9.97CMKK121 pKa = 10.21EE122 pKa = 3.95DD123 pKa = 3.97TRR125 pKa = 11.84DD126 pKa = 3.83GPDD129 pKa = 3.42GGPFEE134 pKa = 4.72YY135 pKa = 8.77GTYY138 pKa = 10.75SKK140 pKa = 9.96TPGNGQGKK148 pKa = 10.3RR149 pKa = 11.84NDD151 pKa = 3.3LTDD154 pKa = 3.44AVEE157 pKa = 4.0ALEE160 pKa = 4.42AGGVEE165 pKa = 4.33AVVSKK170 pKa = 11.01HH171 pKa = 5.41PNTYY175 pKa = 9.13VRR177 pKa = 11.84YY178 pKa = 9.29HH179 pKa = 6.61RR180 pKa = 11.84GIHH183 pKa = 6.31ALYY186 pKa = 9.48QAKK189 pKa = 10.52LEE191 pKa = 4.95AIASRR196 pKa = 11.84QQRR199 pKa = 11.84NVKK202 pKa = 8.46TAVFYY207 pKa = 11.11GEE209 pKa = 4.42PGTGKK214 pKa = 6.71THH216 pKa = 7.2CAFEE220 pKa = 4.16LARR223 pKa = 11.84ITGEE227 pKa = 3.84EE228 pKa = 4.22IYY230 pKa = 10.59ILNAPAGRR238 pKa = 11.84NQSVWFNGYY247 pKa = 8.23QNQKK251 pKa = 9.73ILVVDD256 pKa = 4.13EE257 pKa = 4.97MNGDD261 pKa = 3.97WIGWQLLLRR270 pKa = 11.84MTDD273 pKa = 4.21KK274 pKa = 11.41YY275 pKa = 11.03PLQCQTKK282 pKa = 10.07GGMVWAMWEE291 pKa = 4.04IVIFTSNAHH300 pKa = 5.64WEE302 pKa = 4.02DD303 pKa = 3.36WYY305 pKa = 10.38PYY307 pKa = 9.7YY308 pKa = 10.98AGGMDD313 pKa = 3.31KK314 pKa = 10.96AALKK318 pKa = 10.47RR319 pKa = 11.84RR320 pKa = 11.84IHH322 pKa = 6.1KK323 pKa = 9.25VVKK326 pKa = 10.17FYY328 pKa = 11.25GSEE331 pKa = 3.78ASGNFTKK338 pKa = 10.33KK339 pKa = 10.74YY340 pKa = 9.53YY341 pKa = 10.91DD342 pKa = 4.07PDD344 pKa = 3.64LPEE347 pKa = 5.21ANWTVATTDD356 pKa = 3.37VIATFLRR363 pKa = 11.84NGTVRR368 pKa = 11.84PHH370 pKa = 5.92TDD372 pKa = 2.99EE373 pKa = 4.26EE374 pKa = 4.71LEE376 pKa = 4.15EE377 pKa = 4.41VEE379 pKa = 4.33DD380 pKa = 4.2VVMNTQQEE388 pKa = 4.58SQGTQTTEE396 pKa = 3.79LVEE399 pKa = 5.35DD400 pKa = 4.42SDD402 pKa = 6.05DD403 pKa = 3.84EE404 pKa = 4.21MSEE407 pKa = 4.29TFF409 pKa = 4.24
Molecular weight: 46.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6GIH8|C6GIH8_9VIRU Coat protein OS=Circovirus-like genome RW-B OX=642252 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TNPYY8 pKa = 9.39QRR10 pKa = 11.84ASLPPKK16 pKa = 9.38KK17 pKa = 9.86RR18 pKa = 11.84YY19 pKa = 9.42RR20 pKa = 11.84ADD22 pKa = 3.35QQPAIPRR29 pKa = 11.84LRR31 pKa = 11.84LPYY34 pKa = 10.26HH35 pKa = 6.73LGGPLYY41 pKa = 10.52VSRR44 pKa = 11.84PLPMARR50 pKa = 11.84RR51 pKa = 11.84GAPTGRR57 pKa = 11.84YY58 pKa = 9.15ASNPRR63 pKa = 11.84MGEE66 pKa = 4.05LKK68 pKa = 9.9TVDD71 pKa = 3.78YY72 pKa = 10.72QFSAAYY78 pKa = 8.95AVPYY82 pKa = 10.67VGDD85 pKa = 3.85SQPFPLLNCTNATGNIQCVNLVQQGTGVAQRR116 pKa = 11.84IGNKK120 pKa = 8.72ICMRR124 pKa = 11.84SLRR127 pKa = 11.84IRR129 pKa = 11.84LRR131 pKa = 11.84FVEE134 pKa = 5.53AGTDD138 pKa = 3.8DD139 pKa = 3.62NQAPYY144 pKa = 9.69FARR147 pKa = 11.84IMLIYY152 pKa = 10.39DD153 pKa = 4.04RR154 pKa = 11.84NPNSLYY160 pKa = 10.33IATNNILANALQNNTLGNGTLDD182 pKa = 3.16SSLNPTYY189 pKa = 10.58YY190 pKa = 10.78DD191 pKa = 3.07RR192 pKa = 11.84FVVLRR197 pKa = 11.84DD198 pKa = 3.44WYY200 pKa = 9.09QTIPTFQEE208 pKa = 3.91STSNVAPNWIIDD220 pKa = 3.73DD221 pKa = 4.11FVNLKK226 pKa = 10.23NLEE229 pKa = 4.02TTYY232 pKa = 11.28NGTANPMTINQVSVGALLLFVISDD256 pKa = 3.59QVAATSDD263 pKa = 3.48AVALGGNLRR272 pKa = 11.84LRR274 pKa = 11.84FHH276 pKa = 7.04DD277 pKa = 3.49QQ278 pKa = 2.85
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TNPYY8 pKa = 9.39QRR10 pKa = 11.84ASLPPKK16 pKa = 9.38KK17 pKa = 9.86RR18 pKa = 11.84YY19 pKa = 9.42RR20 pKa = 11.84ADD22 pKa = 3.35QQPAIPRR29 pKa = 11.84LRR31 pKa = 11.84LPYY34 pKa = 10.26HH35 pKa = 6.73LGGPLYY41 pKa = 10.52VSRR44 pKa = 11.84PLPMARR50 pKa = 11.84RR51 pKa = 11.84GAPTGRR57 pKa = 11.84YY58 pKa = 9.15ASNPRR63 pKa = 11.84MGEE66 pKa = 4.05LKK68 pKa = 9.9TVDD71 pKa = 3.78YY72 pKa = 10.72QFSAAYY78 pKa = 8.95AVPYY82 pKa = 10.67VGDD85 pKa = 3.85SQPFPLLNCTNATGNIQCVNLVQQGTGVAQRR116 pKa = 11.84IGNKK120 pKa = 8.72ICMRR124 pKa = 11.84SLRR127 pKa = 11.84IRR129 pKa = 11.84LRR131 pKa = 11.84FVEE134 pKa = 5.53AGTDD138 pKa = 3.8DD139 pKa = 3.62NQAPYY144 pKa = 9.69FARR147 pKa = 11.84IMLIYY152 pKa = 10.39DD153 pKa = 4.04RR154 pKa = 11.84NPNSLYY160 pKa = 10.33IATNNILANALQNNTLGNGTLDD182 pKa = 3.16SSLNPTYY189 pKa = 10.58YY190 pKa = 10.78DD191 pKa = 3.07RR192 pKa = 11.84FVVLRR197 pKa = 11.84DD198 pKa = 3.44WYY200 pKa = 9.09QTIPTFQEE208 pKa = 3.91STSNVAPNWIIDD220 pKa = 3.73DD221 pKa = 4.11FVNLKK226 pKa = 10.23NLEE229 pKa = 4.02TTYY232 pKa = 11.28NGTANPMTINQVSVGALLLFVISDD256 pKa = 3.59QVAATSDD263 pKa = 3.48AVALGGNLRR272 pKa = 11.84LRR274 pKa = 11.84FHH276 pKa = 7.04DD277 pKa = 3.49QQ278 pKa = 2.85
Molecular weight: 31.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
687 |
278 |
409 |
343.5 |
38.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.715 ± 0.588 | 1.164 ± 0.055 |
5.24 ± 0.131 | 5.822 ± 2.806 |
3.202 ± 0.022 | 6.987 ± 0.558 |
1.601 ± 0.564 | 3.93 ± 0.708 |
3.493 ± 0.855 | 6.987 ± 1.975 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.62 ± 0.296 | 6.55 ± 1.563 |
5.386 ± 1.158 | 5.24 ± 0.33 |
7.132 ± 0.5 | 5.095 ± 0.193 |
7.569 ± 0.47 | 7.569 ± 0.931 |
1.747 ± 0.658 | 4.949 ± 0.286 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
