
Beauveria bassiana (White muscardine disease fungus) (Tritirachium shiotae)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta;
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
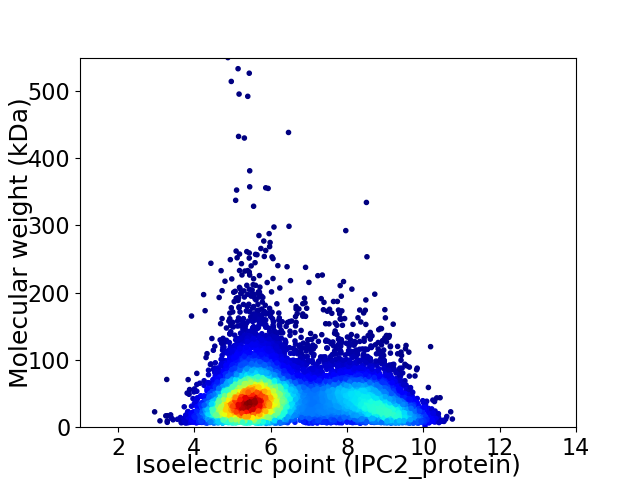
Virtual 2D-PAGE plot for 10656 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
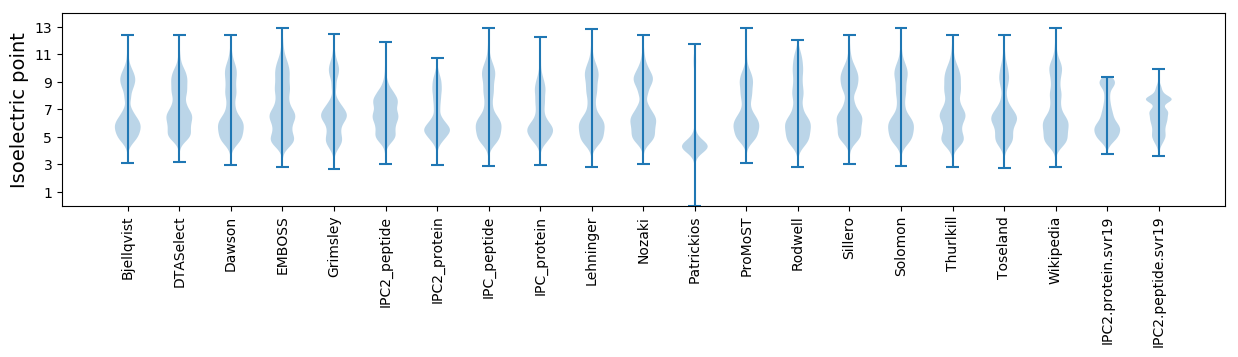
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N6NWK6|A0A2N6NWK6_BEABA Uncharacterized protein OS=Beauveria bassiana OX=176275 GN=BM221_001757 PE=4 SV=1
MM1 pKa = 7.32SNYY4 pKa = 10.16NGRR7 pKa = 11.84HH8 pKa = 5.09GPNVANYY15 pKa = 10.07LRR17 pKa = 11.84DD18 pKa = 3.73LNTISPQEE26 pKa = 3.9GSVDD30 pKa = 3.38EE31 pKa = 4.36NFNIEE36 pKa = 3.76GDD38 pKa = 3.58LALFTNTEE46 pKa = 3.91FFDD49 pKa = 4.93FEE51 pKa = 4.58TGQNTDD57 pKa = 3.67FQAQPVKK64 pKa = 10.15EE65 pKa = 4.36ATHH68 pKa = 5.75STPSSDD74 pKa = 4.38DD75 pKa = 3.21VSAAPSVMGDD85 pKa = 3.4MPNLDD90 pKa = 5.16FISSTYY96 pKa = 10.27IFFYY100 pKa = 10.89VLFYY104 pKa = 10.61FPP106 pKa = 5.4
MM1 pKa = 7.32SNYY4 pKa = 10.16NGRR7 pKa = 11.84HH8 pKa = 5.09GPNVANYY15 pKa = 10.07LRR17 pKa = 11.84DD18 pKa = 3.73LNTISPQEE26 pKa = 3.9GSVDD30 pKa = 3.38EE31 pKa = 4.36NFNIEE36 pKa = 3.76GDD38 pKa = 3.58LALFTNTEE46 pKa = 3.91FFDD49 pKa = 4.93FEE51 pKa = 4.58TGQNTDD57 pKa = 3.67FQAQPVKK64 pKa = 10.15EE65 pKa = 4.36ATHH68 pKa = 5.75STPSSDD74 pKa = 4.38DD75 pKa = 3.21VSAAPSVMGDD85 pKa = 3.4MPNLDD90 pKa = 5.16FISSTYY96 pKa = 10.27IFFYY100 pKa = 10.89VLFYY104 pKa = 10.61FPP106 pKa = 5.4
Molecular weight: 11.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N6NLC1|A0A2N6NLC1_BEABA Cation-transporting ATPase OS=Beauveria bassiana OX=176275 GN=BM221_006212 PE=3 SV=1
MM1 pKa = 7.27SAVKK5 pKa = 10.14AAWTPYY11 pKa = 9.16SQSASSPPPPAPPAPSPSSQHH32 pKa = 4.95SASRR36 pKa = 11.84IAAMEE41 pKa = 3.94AVAPAHH47 pKa = 6.5RR48 pKa = 11.84AGSAHH53 pKa = 6.74AADD56 pKa = 4.26TASVPATGGGRR67 pKa = 11.84CSSSTKK73 pKa = 10.32LLYY76 pKa = 10.13VRR78 pKa = 11.84CSVALHH84 pKa = 7.04DD85 pKa = 4.15GAARR89 pKa = 11.84PRR91 pKa = 11.84VAADD95 pKa = 3.43GLSAGGSCTRR105 pKa = 11.84KK106 pKa = 9.56AARR109 pKa = 11.84ASSARR114 pKa = 11.84RR115 pKa = 11.84DD116 pKa = 3.27AATRR120 pKa = 11.84SCVEE124 pKa = 3.71EE125 pKa = 4.89RR126 pKa = 11.84SAQRR130 pKa = 11.84DD131 pKa = 3.22RR132 pKa = 11.84MAEE135 pKa = 3.61AKK137 pKa = 8.9MAQSLACCMAKK148 pKa = 8.95STWAIHH154 pKa = 5.16QSLVHH159 pKa = 6.01SRR161 pKa = 11.84RR162 pKa = 11.84CTGSILEE169 pKa = 4.62CVASRR174 pKa = 11.84CMPWPMAMRR183 pKa = 11.84IAKK186 pKa = 9.16PIQCHH191 pKa = 5.4AFATISSCPSEE202 pKa = 3.93YY203 pKa = 10.4HH204 pKa = 6.35VISSATCTYY213 pKa = 9.32MHH215 pKa = 6.49MHH217 pKa = 7.24AHH219 pKa = 4.58QQHH222 pKa = 7.14RR223 pKa = 11.84IQQRR227 pKa = 11.84QHH229 pKa = 4.7EE230 pKa = 4.61RR231 pKa = 11.84VPRR234 pKa = 11.84VAAAVLRR241 pKa = 11.84HH242 pKa = 5.7KK243 pKa = 10.89VLLVHH248 pKa = 6.25NRR250 pKa = 11.84PSRR253 pKa = 11.84VHH255 pKa = 5.46AVKK258 pKa = 10.8VGAQQAALEE267 pKa = 4.23RR268 pKa = 11.84RR269 pKa = 11.84GAPRR273 pKa = 11.84QGLGRR278 pKa = 11.84HH279 pKa = 5.63ALQLVRR285 pKa = 11.84RR286 pKa = 11.84VAPRR290 pKa = 11.84KK291 pKa = 9.17LAAAKK296 pKa = 10.04VKK298 pKa = 10.44GVASLRR304 pKa = 11.84LRR306 pKa = 11.84DD307 pKa = 4.59LPHH310 pKa = 5.87QVAKK314 pKa = 10.15VGHH317 pKa = 5.53VAADD321 pKa = 3.56EE322 pKa = 4.01AHH324 pKa = 6.43AAEE327 pKa = 4.53LPEE330 pKa = 5.16LGVAQVLPVDD340 pKa = 3.95EE341 pKa = 4.72VGQVAGLMFLVLCC354 pKa = 5.17
MM1 pKa = 7.27SAVKK5 pKa = 10.14AAWTPYY11 pKa = 9.16SQSASSPPPPAPPAPSPSSQHH32 pKa = 4.95SASRR36 pKa = 11.84IAAMEE41 pKa = 3.94AVAPAHH47 pKa = 6.5RR48 pKa = 11.84AGSAHH53 pKa = 6.74AADD56 pKa = 4.26TASVPATGGGRR67 pKa = 11.84CSSSTKK73 pKa = 10.32LLYY76 pKa = 10.13VRR78 pKa = 11.84CSVALHH84 pKa = 7.04DD85 pKa = 4.15GAARR89 pKa = 11.84PRR91 pKa = 11.84VAADD95 pKa = 3.43GLSAGGSCTRR105 pKa = 11.84KK106 pKa = 9.56AARR109 pKa = 11.84ASSARR114 pKa = 11.84RR115 pKa = 11.84DD116 pKa = 3.27AATRR120 pKa = 11.84SCVEE124 pKa = 3.71EE125 pKa = 4.89RR126 pKa = 11.84SAQRR130 pKa = 11.84DD131 pKa = 3.22RR132 pKa = 11.84MAEE135 pKa = 3.61AKK137 pKa = 8.9MAQSLACCMAKK148 pKa = 8.95STWAIHH154 pKa = 5.16QSLVHH159 pKa = 6.01SRR161 pKa = 11.84RR162 pKa = 11.84CTGSILEE169 pKa = 4.62CVASRR174 pKa = 11.84CMPWPMAMRR183 pKa = 11.84IAKK186 pKa = 9.16PIQCHH191 pKa = 5.4AFATISSCPSEE202 pKa = 3.93YY203 pKa = 10.4HH204 pKa = 6.35VISSATCTYY213 pKa = 9.32MHH215 pKa = 6.49MHH217 pKa = 7.24AHH219 pKa = 4.58QQHH222 pKa = 7.14RR223 pKa = 11.84IQQRR227 pKa = 11.84QHH229 pKa = 4.7EE230 pKa = 4.61RR231 pKa = 11.84VPRR234 pKa = 11.84VAAAVLRR241 pKa = 11.84HH242 pKa = 5.7KK243 pKa = 10.89VLLVHH248 pKa = 6.25NRR250 pKa = 11.84PSRR253 pKa = 11.84VHH255 pKa = 5.46AVKK258 pKa = 10.8VGAQQAALEE267 pKa = 4.23RR268 pKa = 11.84RR269 pKa = 11.84GAPRR273 pKa = 11.84QGLGRR278 pKa = 11.84HH279 pKa = 5.63ALQLVRR285 pKa = 11.84RR286 pKa = 11.84VAPRR290 pKa = 11.84KK291 pKa = 9.17LAAAKK296 pKa = 10.04VKK298 pKa = 10.44GVASLRR304 pKa = 11.84LRR306 pKa = 11.84DD307 pKa = 4.59LPHH310 pKa = 5.87QVAKK314 pKa = 10.15VGHH317 pKa = 5.53VAADD321 pKa = 3.56EE322 pKa = 4.01AHH324 pKa = 6.43AAEE327 pKa = 4.53LPEE330 pKa = 5.16LGVAQVLPVDD340 pKa = 3.95EE341 pKa = 4.72VGQVAGLMFLVLCC354 pKa = 5.17
Molecular weight: 37.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4698728 |
19 |
4917 |
440.9 |
48.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.732 ± 0.025 | 1.223 ± 0.008 |
5.93 ± 0.017 | 5.937 ± 0.021 |
3.579 ± 0.016 | 7.043 ± 0.025 |
2.409 ± 0.012 | 4.58 ± 0.018 |
4.7 ± 0.021 | 8.89 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.244 ± 0.009 | 3.482 ± 0.013 |
5.812 ± 0.026 | 4.027 ± 0.017 |
6.307 ± 0.024 | 7.936 ± 0.03 |
5.841 ± 0.017 | 6.293 ± 0.018 |
1.423 ± 0.008 | 2.612 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
