
Aureococcus anophagefferens (Harmful bloom alga)
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Ochrophyta; Pelagophyceae; Pelagomonadales; Aureococcus
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
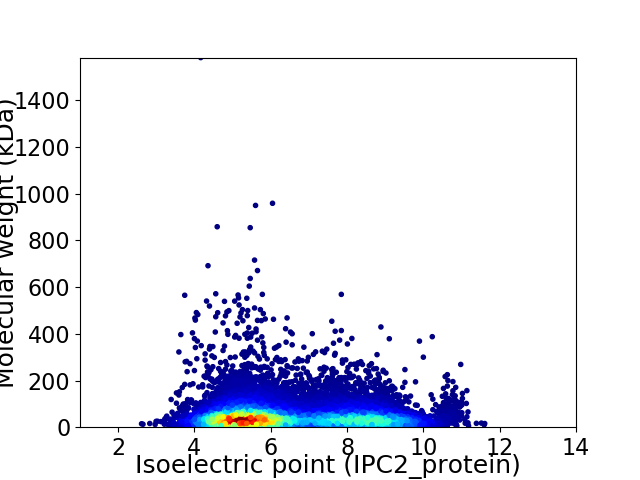
Virtual 2D-PAGE plot for 11501 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F0YB55|F0YB55_AURAN AAA_12 domain-containing protein (Fragment) OS=Aureococcus anophagefferens OX=44056 GN=AURANDRAFT_17677 PE=4 SV=1
MM1 pKa = 7.42GLHH4 pKa = 6.51RR5 pKa = 11.84ALAALLVLALGAAASEE21 pKa = 4.82DD22 pKa = 3.73FSQVAKK28 pKa = 10.71LVASDD33 pKa = 4.15GAAGDD38 pKa = 3.67EE39 pKa = 4.76FGWSVASSGDD49 pKa = 3.58FVVVGAYY56 pKa = 10.56LDD58 pKa = 4.91DD59 pKa = 5.06DD60 pKa = 5.11AGTSSGSAYY69 pKa = 10.43VFRR72 pKa = 11.84TVDD75 pKa = 3.21GVTEE79 pKa = 4.12QIAKK83 pKa = 10.12LVSSDD88 pKa = 4.77AAAQDD93 pKa = 3.4QFGASVAIHH102 pKa = 6.49NDD104 pKa = 3.11IIVVGANHH112 pKa = 7.46DD113 pKa = 3.89DD114 pKa = 3.67DD115 pKa = 6.59AGYY118 pKa = 10.97DD119 pKa = 3.44SGSAYY124 pKa = 9.7IFKK127 pKa = 10.19TSDD130 pKa = 3.59GGATWPQVAKK140 pKa = 10.68LVASDD145 pKa = 4.15GAAGDD150 pKa = 3.68EE151 pKa = 4.68FGWSIASSGDD161 pKa = 3.48FVVVGANLDD170 pKa = 3.85DD171 pKa = 4.96DD172 pKa = 5.63AGTSSGSAYY181 pKa = 10.43VFRR184 pKa = 11.84TVDD187 pKa = 3.21GVTEE191 pKa = 4.12QIAKK195 pKa = 10.12LVSSDD200 pKa = 4.77AAAQDD205 pKa = 3.45QFGVSVAIHH214 pKa = 6.37NDD216 pKa = 3.16IIVVGAAWNDD226 pKa = 3.7DD227 pKa = 3.53AGSNSGSAYY236 pKa = 9.75IFKK239 pKa = 10.19TSDD242 pKa = 3.59GGATWPQVAKK252 pKa = 10.65LVADD256 pKa = 3.96DD257 pKa = 4.68AEE259 pKa = 5.53AYY261 pKa = 10.36DD262 pKa = 3.79LFGWSVAVEE271 pKa = 4.18GNLVVVGALQDD282 pKa = 3.6DD283 pKa = 4.55TNAGSAYY290 pKa = 10.47AFRR293 pKa = 11.84TTDD296 pKa = 4.46GGATWTQAAKK306 pKa = 10.23LAPGSAAYY314 pKa = 10.19HH315 pKa = 6.56FGTSLSISGNTLAVGAYY332 pKa = 7.14EE333 pKa = 4.4ANSFAGGAYY342 pKa = 10.5LFSTTDD348 pKa = 3.41NGASWTQDD356 pKa = 2.59AYY358 pKa = 11.42LQASDD363 pKa = 4.09YY364 pKa = 11.49ASADD368 pKa = 3.38RR369 pKa = 11.84FYY371 pKa = 11.2EE372 pKa = 4.0ALVLSGNRR380 pKa = 11.84LVVGATYY387 pKa = 10.99DD388 pKa = 3.27NDD390 pKa = 3.43MGGNSGSVYY399 pKa = 10.09IFEE402 pKa = 5.1APQPAPSPP410 pKa = 3.81
MM1 pKa = 7.42GLHH4 pKa = 6.51RR5 pKa = 11.84ALAALLVLALGAAASEE21 pKa = 4.82DD22 pKa = 3.73FSQVAKK28 pKa = 10.71LVASDD33 pKa = 4.15GAAGDD38 pKa = 3.67EE39 pKa = 4.76FGWSVASSGDD49 pKa = 3.58FVVVGAYY56 pKa = 10.56LDD58 pKa = 4.91DD59 pKa = 5.06DD60 pKa = 5.11AGTSSGSAYY69 pKa = 10.43VFRR72 pKa = 11.84TVDD75 pKa = 3.21GVTEE79 pKa = 4.12QIAKK83 pKa = 10.12LVSSDD88 pKa = 4.77AAAQDD93 pKa = 3.4QFGASVAIHH102 pKa = 6.49NDD104 pKa = 3.11IIVVGANHH112 pKa = 7.46DD113 pKa = 3.89DD114 pKa = 3.67DD115 pKa = 6.59AGYY118 pKa = 10.97DD119 pKa = 3.44SGSAYY124 pKa = 9.7IFKK127 pKa = 10.19TSDD130 pKa = 3.59GGATWPQVAKK140 pKa = 10.68LVASDD145 pKa = 4.15GAAGDD150 pKa = 3.68EE151 pKa = 4.68FGWSIASSGDD161 pKa = 3.48FVVVGANLDD170 pKa = 3.85DD171 pKa = 4.96DD172 pKa = 5.63AGTSSGSAYY181 pKa = 10.43VFRR184 pKa = 11.84TVDD187 pKa = 3.21GVTEE191 pKa = 4.12QIAKK195 pKa = 10.12LVSSDD200 pKa = 4.77AAAQDD205 pKa = 3.45QFGVSVAIHH214 pKa = 6.37NDD216 pKa = 3.16IIVVGAAWNDD226 pKa = 3.7DD227 pKa = 3.53AGSNSGSAYY236 pKa = 9.75IFKK239 pKa = 10.19TSDD242 pKa = 3.59GGATWPQVAKK252 pKa = 10.65LVADD256 pKa = 3.96DD257 pKa = 4.68AEE259 pKa = 5.53AYY261 pKa = 10.36DD262 pKa = 3.79LFGWSVAVEE271 pKa = 4.18GNLVVVGALQDD282 pKa = 3.6DD283 pKa = 4.55TNAGSAYY290 pKa = 10.47AFRR293 pKa = 11.84TTDD296 pKa = 4.46GGATWTQAAKK306 pKa = 10.23LAPGSAAYY314 pKa = 10.19HH315 pKa = 6.56FGTSLSISGNTLAVGAYY332 pKa = 7.14EE333 pKa = 4.4ANSFAGGAYY342 pKa = 10.5LFSTTDD348 pKa = 3.41NGASWTQDD356 pKa = 2.59AYY358 pKa = 11.42LQASDD363 pKa = 4.09YY364 pKa = 11.49ASADD368 pKa = 3.38RR369 pKa = 11.84FYY371 pKa = 11.2EE372 pKa = 4.0ALVLSGNRR380 pKa = 11.84LVVGATYY387 pKa = 10.99DD388 pKa = 3.27NDD390 pKa = 3.43MGGNSGSVYY399 pKa = 10.09IFEE402 pKa = 5.1APQPAPSPP410 pKa = 3.81
Molecular weight: 41.62 kDa
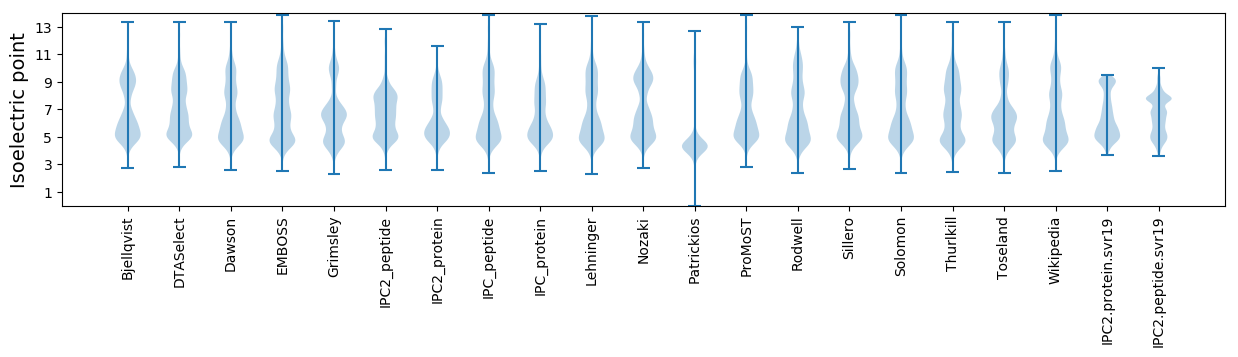
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F0YT44|F0YT44_AURAN Expressed protein OS=Aureococcus anophagefferens OX=44056 GN=AURANDRAFT_69566 PE=4 SV=1
RR1 pKa = 6.84PTSSSRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84SSARR13 pKa = 11.84APTPARR19 pKa = 11.84GRR21 pKa = 11.84RR22 pKa = 11.84TTTPRR27 pKa = 11.84TTTPRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84PTRR38 pKa = 11.84PRR40 pKa = 11.84SRR42 pKa = 11.84SGPTASRR49 pKa = 11.84TTRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84PTATSPAAPRR64 pKa = 11.84TRR66 pKa = 11.84TGSTSATRR74 pKa = 11.84RR75 pKa = 11.84AWRR78 pKa = 11.84RR79 pKa = 11.84STSPRR84 pKa = 11.84PSRR87 pKa = 11.84PRR89 pKa = 11.84SPRR92 pKa = 11.84TRR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84GAAGSRR106 pKa = 11.84SRR108 pKa = 11.84SPRR111 pKa = 11.84PSAAPRR117 pKa = 11.84RR118 pKa = 11.84PANRR122 pKa = 11.84GRR124 pKa = 11.84GATTNRR130 pKa = 11.84GRR132 pKa = 11.84GATTTRR138 pKa = 11.84PSASPCKK145 pKa = 9.9CHH147 pKa = 7.25LVV149 pKa = 3.46
RR1 pKa = 6.84PTSSSRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84SSARR13 pKa = 11.84APTPARR19 pKa = 11.84GRR21 pKa = 11.84RR22 pKa = 11.84TTTPRR27 pKa = 11.84TTTPRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84PTRR38 pKa = 11.84PRR40 pKa = 11.84SRR42 pKa = 11.84SGPTASRR49 pKa = 11.84TTRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84PTATSPAAPRR64 pKa = 11.84TRR66 pKa = 11.84TGSTSATRR74 pKa = 11.84RR75 pKa = 11.84AWRR78 pKa = 11.84RR79 pKa = 11.84STSPRR84 pKa = 11.84PSRR87 pKa = 11.84PRR89 pKa = 11.84SPRR92 pKa = 11.84TRR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84GAAGSRR106 pKa = 11.84SRR108 pKa = 11.84SPRR111 pKa = 11.84PSAAPRR117 pKa = 11.84RR118 pKa = 11.84PANRR122 pKa = 11.84GRR124 pKa = 11.84GATTNRR130 pKa = 11.84GRR132 pKa = 11.84GATTTRR138 pKa = 11.84PSASPCKK145 pKa = 9.9CHH147 pKa = 7.25LVV149 pKa = 3.46
Molecular weight: 16.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6025983 |
29 |
15048 |
524.0 |
56.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.294 ± 0.056 | 1.833 ± 0.012 |
6.889 ± 0.024 | 5.616 ± 0.023 |
3.333 ± 0.014 | 8.227 ± 0.03 |
2.094 ± 0.011 | 2.571 ± 0.018 |
3.71 ± 0.024 | 9.232 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.592 ± 0.011 | 2.148 ± 0.013 |
5.84 ± 0.023 | 2.295 ± 0.013 |
8.156 ± 0.048 | 5.725 ± 0.025 |
4.551 ± 0.021 | 7.289 ± 0.022 |
1.342 ± 0.008 | 2.263 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
