
Arthrobacter phage Idaho
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
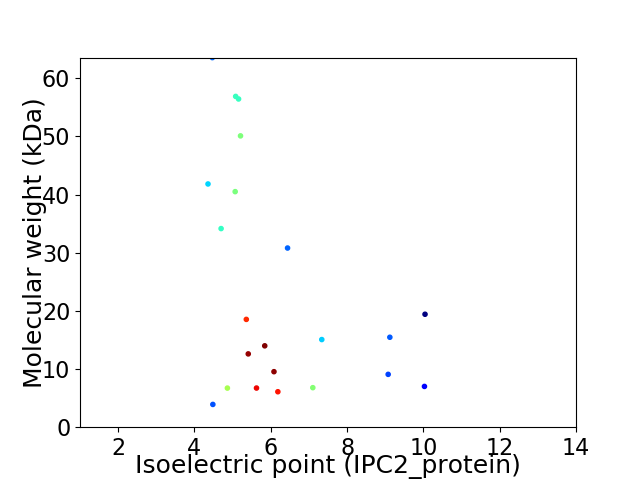
Virtual 2D-PAGE plot for 22 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D6T834|A0A4D6T834_9CAUD Uncharacterized protein OS=Arthrobacter phage Idaho OX=2565509 GN=1 PE=4 SV=1
MM1 pKa = 7.84PNLTDD6 pKa = 5.65LLTLTKK12 pKa = 9.82RR13 pKa = 11.84AIMGSPSPLEE23 pKa = 4.39FGSSTSGGYY32 pKa = 7.41TWADD36 pKa = 3.02GMALSPILPPSIAGGAVSVKK56 pKa = 10.43DD57 pKa = 3.98ALRR60 pKa = 11.84CPPVARR66 pKa = 11.84GVGLYY71 pKa = 9.71AAAMLQMDD79 pKa = 4.98WEE81 pKa = 5.62AGDD84 pKa = 3.89ATKK87 pKa = 10.7AWLNKK92 pKa = 9.8GSGIVSPRR100 pKa = 11.84LRR102 pKa = 11.84NVRR105 pKa = 11.84TMTDD109 pKa = 3.0LLLANRR115 pKa = 11.84ALWITSGTGYY125 pKa = 10.73DD126 pKa = 3.35RR127 pKa = 11.84QEE129 pKa = 4.02AVHH132 pKa = 6.29LHH134 pKa = 4.91AHH136 pKa = 6.39LWSVDD141 pKa = 3.48EE142 pKa = 5.37LGTISLSGVLPDD154 pKa = 4.37ANGDD158 pKa = 3.72YY159 pKa = 11.55VMGGRR164 pKa = 11.84VVTNVANLIYY174 pKa = 10.5FPGAMPLGFLEE185 pKa = 4.24NAQDD189 pKa = 3.64SLSYY193 pKa = 10.96YY194 pKa = 10.76LDD196 pKa = 3.06ILATIKK202 pKa = 10.62SRR204 pKa = 11.84AAAPMAVMEE213 pKa = 4.5LKK215 pKa = 10.47ILDD218 pKa = 4.36SYY220 pKa = 11.5DD221 pKa = 3.44GPEE224 pKa = 4.85PGDD227 pKa = 3.7DD228 pKa = 5.72DD229 pKa = 4.85YY230 pKa = 12.07DD231 pKa = 4.08EE232 pKa = 6.68DD233 pKa = 4.66EE234 pKa = 4.81DD235 pKa = 6.04PIRR238 pKa = 11.84QTQAAYY244 pKa = 9.58VAARR248 pKa = 11.84RR249 pKa = 11.84MPDD252 pKa = 2.89GAVTVTPKK260 pKa = 10.82DD261 pKa = 2.96VDD263 pKa = 4.89LIVHH267 pKa = 5.09QVEE270 pKa = 4.92DD271 pKa = 4.54DD272 pKa = 3.32GHH274 pKa = 5.79MLIEE278 pKa = 4.05ARR280 pKa = 11.84AALRR284 pKa = 11.84VDD286 pKa = 3.86VANHH290 pKa = 6.05LNMPASMLDD299 pKa = 3.48GEE301 pKa = 4.97TGSSSDD307 pKa = 4.35YY308 pKa = 11.59ANTLQKK314 pKa = 10.69RR315 pKa = 11.84NEE317 pKa = 4.03FEE319 pKa = 4.47DD320 pKa = 4.02LSLALFTDD328 pKa = 5.68PIAARR333 pKa = 11.84LSMDD337 pKa = 3.79DD338 pKa = 3.33VTPPGEE344 pKa = 4.1LVDD347 pKa = 5.83FKK349 pKa = 10.41PLDD352 pKa = 3.7MAPVAVGNTGDD363 pKa = 4.3AVGNLQAGQLPRR375 pKa = 11.84DD376 pKa = 3.65NKK378 pKa = 10.3NRR380 pKa = 11.84EE381 pKa = 3.75ITRR384 pKa = 11.84DD385 pKa = 3.65PEE387 pKa = 3.85NALL390 pKa = 3.63
MM1 pKa = 7.84PNLTDD6 pKa = 5.65LLTLTKK12 pKa = 9.82RR13 pKa = 11.84AIMGSPSPLEE23 pKa = 4.39FGSSTSGGYY32 pKa = 7.41TWADD36 pKa = 3.02GMALSPILPPSIAGGAVSVKK56 pKa = 10.43DD57 pKa = 3.98ALRR60 pKa = 11.84CPPVARR66 pKa = 11.84GVGLYY71 pKa = 9.71AAAMLQMDD79 pKa = 4.98WEE81 pKa = 5.62AGDD84 pKa = 3.89ATKK87 pKa = 10.7AWLNKK92 pKa = 9.8GSGIVSPRR100 pKa = 11.84LRR102 pKa = 11.84NVRR105 pKa = 11.84TMTDD109 pKa = 3.0LLLANRR115 pKa = 11.84ALWITSGTGYY125 pKa = 10.73DD126 pKa = 3.35RR127 pKa = 11.84QEE129 pKa = 4.02AVHH132 pKa = 6.29LHH134 pKa = 4.91AHH136 pKa = 6.39LWSVDD141 pKa = 3.48EE142 pKa = 5.37LGTISLSGVLPDD154 pKa = 4.37ANGDD158 pKa = 3.72YY159 pKa = 11.55VMGGRR164 pKa = 11.84VVTNVANLIYY174 pKa = 10.5FPGAMPLGFLEE185 pKa = 4.24NAQDD189 pKa = 3.64SLSYY193 pKa = 10.96YY194 pKa = 10.76LDD196 pKa = 3.06ILATIKK202 pKa = 10.62SRR204 pKa = 11.84AAAPMAVMEE213 pKa = 4.5LKK215 pKa = 10.47ILDD218 pKa = 4.36SYY220 pKa = 11.5DD221 pKa = 3.44GPEE224 pKa = 4.85PGDD227 pKa = 3.7DD228 pKa = 5.72DD229 pKa = 4.85YY230 pKa = 12.07DD231 pKa = 4.08EE232 pKa = 6.68DD233 pKa = 4.66EE234 pKa = 4.81DD235 pKa = 6.04PIRR238 pKa = 11.84QTQAAYY244 pKa = 9.58VAARR248 pKa = 11.84RR249 pKa = 11.84MPDD252 pKa = 2.89GAVTVTPKK260 pKa = 10.82DD261 pKa = 2.96VDD263 pKa = 4.89LIVHH267 pKa = 5.09QVEE270 pKa = 4.92DD271 pKa = 4.54DD272 pKa = 3.32GHH274 pKa = 5.79MLIEE278 pKa = 4.05ARR280 pKa = 11.84AALRR284 pKa = 11.84VDD286 pKa = 3.86VANHH290 pKa = 6.05LNMPASMLDD299 pKa = 3.48GEE301 pKa = 4.97TGSSSDD307 pKa = 4.35YY308 pKa = 11.59ANTLQKK314 pKa = 10.69RR315 pKa = 11.84NEE317 pKa = 4.03FEE319 pKa = 4.47DD320 pKa = 4.02LSLALFTDD328 pKa = 5.68PIAARR333 pKa = 11.84LSMDD337 pKa = 3.79DD338 pKa = 3.33VTPPGEE344 pKa = 4.1LVDD347 pKa = 5.83FKK349 pKa = 10.41PLDD352 pKa = 3.7MAPVAVGNTGDD363 pKa = 4.3AVGNLQAGQLPRR375 pKa = 11.84DD376 pKa = 3.65NKK378 pKa = 10.3NRR380 pKa = 11.84EE381 pKa = 3.75ITRR384 pKa = 11.84DD385 pKa = 3.65PEE387 pKa = 3.85NALL390 pKa = 3.63
Molecular weight: 41.82 kDa
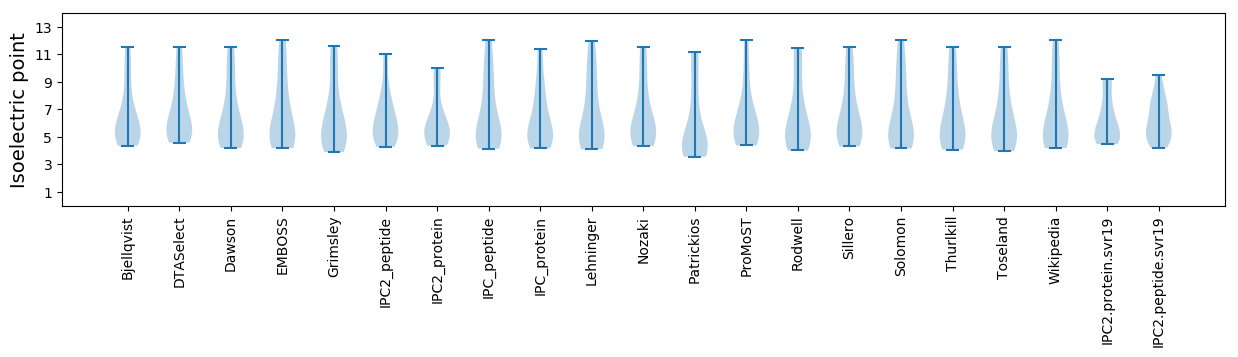
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6TBT1|A0A4D6TBT1_9CAUD HNH endonuclease OS=Arthrobacter phage Idaho OX=2565509 GN=21 PE=4 SV=1
MM1 pKa = 7.48GGSLLTPSAKK11 pKa = 10.01RR12 pKa = 11.84VDD14 pKa = 4.14EE15 pKa = 4.28LRR17 pKa = 11.84AAALALRR24 pKa = 11.84LLPKK28 pKa = 10.58GIRR31 pKa = 11.84NDD33 pKa = 2.95INKK36 pKa = 8.88NRR38 pKa = 11.84RR39 pKa = 11.84AVLNPMWRR47 pKa = 11.84DD48 pKa = 3.11AVNAKK53 pKa = 10.27AVTTMDD59 pKa = 3.83KK60 pKa = 11.05LVLAKK65 pKa = 10.11GARR68 pKa = 11.84VTPGNPARR76 pKa = 11.84AMAATSKK83 pKa = 10.57RR84 pKa = 11.84PLSGGLVPDD93 pKa = 4.8DD94 pKa = 4.59RR95 pKa = 11.84DD96 pKa = 3.42TAVAFEE102 pKa = 4.3FGSPEE107 pKa = 3.72RR108 pKa = 11.84QKK110 pKa = 10.33EE111 pKa = 3.83VEE113 pKa = 4.06YY114 pKa = 10.98ARR116 pKa = 11.84RR117 pKa = 11.84SPKK120 pKa = 10.32GGRR123 pKa = 11.84HH124 pKa = 3.38QVKK127 pKa = 9.68RR128 pKa = 11.84HH129 pKa = 5.18TKK131 pKa = 9.26RR132 pKa = 11.84QLPMLARR139 pKa = 11.84KK140 pKa = 9.68GRR142 pKa = 11.84VIYY145 pKa = 9.78AAWKK149 pKa = 8.06ATAPRR154 pKa = 11.84IIALDD159 pKa = 3.6TQTIARR165 pKa = 11.84LIYY168 pKa = 9.54RR169 pKa = 11.84AYY171 pKa = 9.91EE172 pKa = 4.27GKK174 pKa = 10.8SNN176 pKa = 3.67
MM1 pKa = 7.48GGSLLTPSAKK11 pKa = 10.01RR12 pKa = 11.84VDD14 pKa = 4.14EE15 pKa = 4.28LRR17 pKa = 11.84AAALALRR24 pKa = 11.84LLPKK28 pKa = 10.58GIRR31 pKa = 11.84NDD33 pKa = 2.95INKK36 pKa = 8.88NRR38 pKa = 11.84RR39 pKa = 11.84AVLNPMWRR47 pKa = 11.84DD48 pKa = 3.11AVNAKK53 pKa = 10.27AVTTMDD59 pKa = 3.83KK60 pKa = 11.05LVLAKK65 pKa = 10.11GARR68 pKa = 11.84VTPGNPARR76 pKa = 11.84AMAATSKK83 pKa = 10.57RR84 pKa = 11.84PLSGGLVPDD93 pKa = 4.8DD94 pKa = 4.59RR95 pKa = 11.84DD96 pKa = 3.42TAVAFEE102 pKa = 4.3FGSPEE107 pKa = 3.72RR108 pKa = 11.84QKK110 pKa = 10.33EE111 pKa = 3.83VEE113 pKa = 4.06YY114 pKa = 10.98ARR116 pKa = 11.84RR117 pKa = 11.84SPKK120 pKa = 10.32GGRR123 pKa = 11.84HH124 pKa = 3.38QVKK127 pKa = 9.68RR128 pKa = 11.84HH129 pKa = 5.18TKK131 pKa = 9.26RR132 pKa = 11.84QLPMLARR139 pKa = 11.84KK140 pKa = 9.68GRR142 pKa = 11.84VIYY145 pKa = 9.78AAWKK149 pKa = 8.06ATAPRR154 pKa = 11.84IIALDD159 pKa = 3.6TQTIARR165 pKa = 11.84LIYY168 pKa = 9.54RR169 pKa = 11.84AYY171 pKa = 9.91EE172 pKa = 4.27GKK174 pKa = 10.8SNN176 pKa = 3.67
Molecular weight: 19.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4897 |
35 |
608 |
222.6 |
23.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.825 ± 0.675 | 0.49 ± 0.158 |
7.066 ± 0.506 | 5.228 ± 0.391 |
2.205 ± 0.207 | 8.822 ± 0.292 |
1.348 ± 0.18 | 3.982 ± 0.235 |
4.288 ± 0.514 | 8.863 ± 0.419 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.553 ± 0.225 | 3.206 ± 0.235 |
4.962 ± 0.528 | 3.717 ± 0.268 |
6.106 ± 0.528 | 5.473 ± 0.272 |
6.637 ± 0.376 | 7.106 ± 0.408 |
1.838 ± 0.202 | 2.287 ± 0.283 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
