
Exilibacterium tricleocarpae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Cellvibrionaceae; Exilibacterium
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
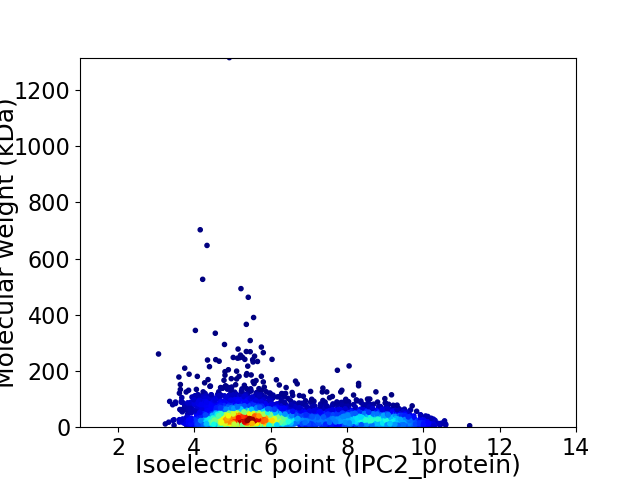
Virtual 2D-PAGE plot for 5556 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A545T5W8|A0A545T5W8_9GAMM HAD-IA family hydrolase OS=Exilibacterium tricleocarpae OX=2591008 GN=FKG94_18135 PE=4 SV=1
MM1 pKa = 7.33LVKK4 pKa = 10.5LRR6 pKa = 11.84QIKK9 pKa = 7.05QHH11 pKa = 6.27KK12 pKa = 9.57NRR14 pKa = 11.84LLKK17 pKa = 10.21RR18 pKa = 11.84WAVLSMLVLLAACDD32 pKa = 4.12DD33 pKa = 4.12NGPAEE38 pKa = 4.4NQPPQAPPLALTTAEE53 pKa = 4.56DD54 pKa = 4.18TPLSITLPGTDD65 pKa = 2.77SDD67 pKa = 4.89GFITYY72 pKa = 9.78HH73 pKa = 5.91SVVTAPVHH81 pKa = 5.06GTLEE85 pKa = 4.4GVAPDD90 pKa = 4.57LLYY93 pKa = 10.53IPDD96 pKa = 3.93ADD98 pKa = 4.14FFGSDD103 pKa = 3.1SFVYY107 pKa = 10.54RR108 pKa = 11.84LTDD111 pKa = 3.13NAGAVSSATVSIEE124 pKa = 3.71VTAVNDD130 pKa = 3.54APVAVDD136 pKa = 4.2DD137 pKa = 4.69SYY139 pKa = 11.99TVAGYY144 pKa = 8.83TSTTIAAPGVLSNDD158 pKa = 3.1SDD160 pKa = 3.72IDD162 pKa = 3.92SASTDD167 pKa = 3.53LTTAAATLDD176 pKa = 3.6TRR178 pKa = 11.84EE179 pKa = 4.3GGTVTMAVDD188 pKa = 3.97GGFTYY193 pKa = 9.99TPPSTVGLTEE203 pKa = 5.01DD204 pKa = 3.3SFEE207 pKa = 4.21YY208 pKa = 10.18TVSDD212 pKa = 3.58NHH214 pKa = 5.95PTAPQSATARR224 pKa = 11.84VTLVLTDD231 pKa = 3.99TGNQPPLARR240 pKa = 11.84DD241 pKa = 3.32LALTTSEE248 pKa = 4.33DD249 pKa = 3.56TSLSILLQGEE259 pKa = 4.24DD260 pKa = 3.33ADD262 pKa = 4.39GVVTAYY268 pKa = 10.77SVITPPTRR276 pKa = 11.84GRR278 pKa = 11.84LSGAAPAMTYY288 pKa = 9.75TPAANYY294 pKa = 9.17FGSDD298 pKa = 3.1SFVYY302 pKa = 10.24GVTDD306 pKa = 3.37NAGATSTATVTITITPVNDD325 pKa = 3.33APVALDD331 pKa = 4.05DD332 pKa = 4.22SFTVARR338 pKa = 11.84NKK340 pKa = 8.68PTVVAAPGVLANDD353 pKa = 3.93SDD355 pKa = 4.32IEE357 pKa = 4.28SAPASLTVTAQTITTARR374 pKa = 11.84GGSVNLDD381 pKa = 3.08SRR383 pKa = 11.84GGFIYY388 pKa = 10.39SPPLTAGVAEE398 pKa = 5.49DD399 pKa = 3.61SFQYY403 pKa = 10.21TLSDD407 pKa = 3.65NDD409 pKa = 3.44PTGARR414 pKa = 11.84FSTGRR419 pKa = 11.84VTLILTGAEE428 pKa = 4.14NQRR431 pKa = 11.84PTAVDD436 pKa = 3.88LNLATPEE443 pKa = 4.03DD444 pKa = 3.92QPLGVTLRR452 pKa = 11.84GEE454 pKa = 4.61DD455 pKa = 3.57GDD457 pKa = 4.43GVIVSYY463 pKa = 11.06AVTVPPTRR471 pKa = 11.84GRR473 pKa = 11.84LTGSAPNLTYY483 pKa = 10.32TPQPDD488 pKa = 3.82YY489 pKa = 11.26FGDD492 pKa = 3.54DD493 pKa = 3.13SFTYY497 pKa = 10.13RR498 pKa = 11.84VTDD501 pKa = 3.77DD502 pKa = 3.44VGATATGTVRR512 pKa = 11.84ITVSTVNDD520 pKa = 3.97APTAAADD527 pKa = 3.47SFNAVRR533 pKa = 11.84NALTTVAAPGVLANDD548 pKa = 3.73SDD550 pKa = 4.24IEE552 pKa = 4.29SAAATLTVTATTLATAQDD570 pKa = 3.96GVVTLNADD578 pKa = 3.36GGFTYY583 pKa = 9.84TPPPTAGLTTDD594 pKa = 3.59SFQYY598 pKa = 10.04TLNDD602 pKa = 3.63NDD604 pKa = 4.33PVDD607 pKa = 4.14PLSATGEE614 pKa = 4.14VTLILTGAEE623 pKa = 4.1NLPPSATDD631 pKa = 3.43LTLTTAEE638 pKa = 4.98DD639 pKa = 3.88SPLSITLQGADD650 pKa = 3.29SDD652 pKa = 4.69GAVATYY658 pKa = 9.36TVTTPPQHH666 pKa = 6.15GRR668 pKa = 11.84LTGVAPDD675 pKa = 3.66LTYY678 pKa = 10.86TPDD681 pKa = 3.45ADD683 pKa = 4.04YY684 pKa = 10.79FGSDD688 pKa = 3.02SFVYY692 pKa = 10.18EE693 pKa = 4.1VTDD696 pKa = 3.89SIGAIASATVTLTVNAVNDD715 pKa = 4.21APTAAADD722 pKa = 3.72SFNAARR728 pKa = 11.84NALTTVAAPGVLANDD743 pKa = 3.91SDD745 pKa = 4.47IEE747 pKa = 4.36STPAALTVTAVTLATAQGGAVTLNADD773 pKa = 3.52GGFTYY778 pKa = 9.84TPPPTAGLTTDD789 pKa = 3.59SFQYY793 pKa = 10.04TLNDD797 pKa = 3.63NDD799 pKa = 4.33PVDD802 pKa = 4.14PLSATGEE809 pKa = 4.14VTLILTGAEE818 pKa = 4.1NLPPSATDD826 pKa = 3.43LTLTTAEE833 pKa = 4.98DD834 pKa = 3.88SPLSITLQGTDD845 pKa = 2.5SDD847 pKa = 4.62GAVVAYY853 pKa = 8.33TVTTPPQHH861 pKa = 5.75GQLAGVAPNLIYY873 pKa = 10.54TPAADD878 pKa = 3.84YY879 pKa = 10.45FGSDD883 pKa = 3.03SFVYY887 pKa = 10.18EE888 pKa = 4.1VTDD891 pKa = 3.89SIGAIASATVTLTINAVNDD910 pKa = 3.84APTAAADD917 pKa = 3.72SFNAARR923 pKa = 11.84NTLTTVAAPGVLANDD938 pKa = 3.76SDD940 pKa = 4.25IEE942 pKa = 4.4SVPAEE947 pKa = 4.21LTVTAATLATAQGGAVTLNADD968 pKa = 3.52GGFTYY973 pKa = 9.81TPPVTTGLSTDD984 pKa = 3.23SFQYY988 pKa = 10.07TLNDD992 pKa = 3.63NDD994 pKa = 4.33PVDD997 pKa = 4.14PLSATGEE1004 pKa = 4.14VTLILTGAEE1013 pKa = 4.1NLPPSATDD1021 pKa = 3.43LTLTTAEE1028 pKa = 4.98DD1029 pKa = 3.88SPLSITLQGADD1040 pKa = 2.91SDD1042 pKa = 4.64GAVVAYY1048 pKa = 8.33TVTTPPQHH1056 pKa = 5.75GQLAGVAPNLTYY1068 pKa = 10.53TPAADD1073 pKa = 3.75YY1074 pKa = 10.39FGSDD1078 pKa = 2.98SFIYY1082 pKa = 10.4AVTDD1086 pKa = 3.47NLGATASATVALTISAVNDD1105 pKa = 3.27APTAVDD1111 pKa = 5.37DD1112 pKa = 5.77DD1113 pKa = 4.32FTAQGNIITLIAAPGVLANDD1133 pKa = 3.98SDD1135 pKa = 4.54VEE1137 pKa = 4.39SADD1140 pKa = 3.78ADD1142 pKa = 3.8LTATAEE1148 pKa = 4.52TIATDD1153 pKa = 3.13QGGSVTINTNGSLLYY1168 pKa = 9.59TPPPAAGVTADD1179 pKa = 3.6SFHH1182 pKa = 6.2YY1183 pKa = 9.41TLNDD1187 pKa = 3.39NDD1189 pKa = 4.26PVDD1192 pKa = 4.08PRR1194 pKa = 11.84TSTGTVTIALTTYY1207 pKa = 10.25TEE1209 pKa = 4.24GLSVQGLPAASGDD1222 pKa = 3.82SATARR1227 pKa = 11.84LYY1229 pKa = 11.27LSGTAEE1235 pKa = 4.69SMLSYY1240 pKa = 10.17QWSADD1245 pKa = 3.48GSWVIDD1251 pKa = 3.4SGQGSEE1257 pKa = 4.91VITLTAPAAAGAANVSVVVEE1277 pKa = 4.25DD1278 pKa = 4.45DD1279 pKa = 3.38SGNRR1283 pKa = 11.84AVASLPLSKK1292 pKa = 10.6AGEE1295 pKa = 4.1TAPVLEE1301 pKa = 4.27SLYY1304 pKa = 11.13LEE1306 pKa = 5.09LPPASAPVEE1315 pKa = 4.21LAVTAYY1321 pKa = 10.65DD1322 pKa = 3.96PDD1324 pKa = 4.27GLALDD1329 pKa = 4.36YY1330 pKa = 9.53TWKK1333 pKa = 10.74SAGATVASGATAQWQPPLPGRR1354 pKa = 11.84YY1355 pKa = 9.44QIDD1358 pKa = 3.46IEE1360 pKa = 4.34VDD1362 pKa = 3.18NSSLVAGGSLQRR1374 pKa = 11.84HH1375 pKa = 4.85YY1376 pKa = 10.23TGGAPWAFFRR1386 pKa = 11.84GSRR1389 pKa = 11.84QGTGVRR1395 pKa = 11.84IPADD1399 pKa = 3.37TSSNSGALKK1408 pKa = 8.28WQRR1411 pKa = 11.84PFTMANCAVADD1422 pKa = 3.81NFVPGVVQGEE1432 pKa = 4.4DD1433 pKa = 3.06GTLYY1437 pKa = 10.73AGSSTEE1443 pKa = 3.77GKK1445 pKa = 10.35LYY1447 pKa = 10.89AFNPDD1452 pKa = 3.68DD1453 pKa = 5.89GVIAWSFTTVGTRR1466 pKa = 11.84IDD1468 pKa = 3.88ATPVIAEE1475 pKa = 4.59DD1476 pKa = 3.7GSIYY1480 pKa = 10.52VVEE1483 pKa = 4.4SGGTVYY1489 pKa = 10.83AVNADD1494 pKa = 3.79GSEE1497 pKa = 3.86KK1498 pKa = 9.64WSYY1501 pKa = 11.39AIGAPVSGSPAIGADD1516 pKa = 3.16GTVYY1520 pKa = 10.84VGTDD1524 pKa = 2.88NGGASVLLALNPADD1538 pKa = 5.72GLPKK1542 pKa = 9.9WSAPFALAAEE1552 pKa = 4.62TRR1554 pKa = 11.84SSVNIGADD1562 pKa = 3.3GTVYY1566 pKa = 10.88ARR1568 pKa = 11.84DD1569 pKa = 3.37FAGNLFAINPADD1581 pKa = 3.68GTQRR1585 pKa = 11.84WQITLGGGSGSTPVIAADD1603 pKa = 3.37GTLYY1607 pKa = 10.62FGSFSPPRR1615 pKa = 11.84LYY1617 pKa = 10.95AVGPDD1622 pKa = 4.75GSPKK1626 pKa = 9.62WDD1628 pKa = 3.13TAMGNPSLAGIGATPSVGADD1648 pKa = 3.01GTVYY1652 pKa = 10.71VGVWEE1657 pKa = 4.64AGLNGSVYY1665 pKa = 10.68AIDD1668 pKa = 3.85PVGGTVVWRR1677 pKa = 11.84RR1678 pKa = 11.84QLDD1681 pKa = 3.73SFVQSAATAVGADD1694 pKa = 3.06GRR1696 pKa = 11.84VYY1698 pKa = 10.22VVSARR1703 pKa = 11.84GVFYY1707 pKa = 10.92ALDD1710 pKa = 4.48PDD1712 pKa = 3.88NGAVDD1717 pKa = 3.38WSFDD1721 pKa = 2.8IGAATNEE1728 pKa = 4.21EE1729 pKa = 4.74SPSPLAIGTDD1739 pKa = 3.54GSIYY1743 pKa = 10.57VYY1745 pKa = 10.26TCDD1748 pKa = 3.12GVLRR1752 pKa = 11.84AFQQ1755 pKa = 3.44
MM1 pKa = 7.33LVKK4 pKa = 10.5LRR6 pKa = 11.84QIKK9 pKa = 7.05QHH11 pKa = 6.27KK12 pKa = 9.57NRR14 pKa = 11.84LLKK17 pKa = 10.21RR18 pKa = 11.84WAVLSMLVLLAACDD32 pKa = 4.12DD33 pKa = 4.12NGPAEE38 pKa = 4.4NQPPQAPPLALTTAEE53 pKa = 4.56DD54 pKa = 4.18TPLSITLPGTDD65 pKa = 2.77SDD67 pKa = 4.89GFITYY72 pKa = 9.78HH73 pKa = 5.91SVVTAPVHH81 pKa = 5.06GTLEE85 pKa = 4.4GVAPDD90 pKa = 4.57LLYY93 pKa = 10.53IPDD96 pKa = 3.93ADD98 pKa = 4.14FFGSDD103 pKa = 3.1SFVYY107 pKa = 10.54RR108 pKa = 11.84LTDD111 pKa = 3.13NAGAVSSATVSIEE124 pKa = 3.71VTAVNDD130 pKa = 3.54APVAVDD136 pKa = 4.2DD137 pKa = 4.69SYY139 pKa = 11.99TVAGYY144 pKa = 8.83TSTTIAAPGVLSNDD158 pKa = 3.1SDD160 pKa = 3.72IDD162 pKa = 3.92SASTDD167 pKa = 3.53LTTAAATLDD176 pKa = 3.6TRR178 pKa = 11.84EE179 pKa = 4.3GGTVTMAVDD188 pKa = 3.97GGFTYY193 pKa = 9.99TPPSTVGLTEE203 pKa = 5.01DD204 pKa = 3.3SFEE207 pKa = 4.21YY208 pKa = 10.18TVSDD212 pKa = 3.58NHH214 pKa = 5.95PTAPQSATARR224 pKa = 11.84VTLVLTDD231 pKa = 3.99TGNQPPLARR240 pKa = 11.84DD241 pKa = 3.32LALTTSEE248 pKa = 4.33DD249 pKa = 3.56TSLSILLQGEE259 pKa = 4.24DD260 pKa = 3.33ADD262 pKa = 4.39GVVTAYY268 pKa = 10.77SVITPPTRR276 pKa = 11.84GRR278 pKa = 11.84LSGAAPAMTYY288 pKa = 9.75TPAANYY294 pKa = 9.17FGSDD298 pKa = 3.1SFVYY302 pKa = 10.24GVTDD306 pKa = 3.37NAGATSTATVTITITPVNDD325 pKa = 3.33APVALDD331 pKa = 4.05DD332 pKa = 4.22SFTVARR338 pKa = 11.84NKK340 pKa = 8.68PTVVAAPGVLANDD353 pKa = 3.93SDD355 pKa = 4.32IEE357 pKa = 4.28SAPASLTVTAQTITTARR374 pKa = 11.84GGSVNLDD381 pKa = 3.08SRR383 pKa = 11.84GGFIYY388 pKa = 10.39SPPLTAGVAEE398 pKa = 5.49DD399 pKa = 3.61SFQYY403 pKa = 10.21TLSDD407 pKa = 3.65NDD409 pKa = 3.44PTGARR414 pKa = 11.84FSTGRR419 pKa = 11.84VTLILTGAEE428 pKa = 4.14NQRR431 pKa = 11.84PTAVDD436 pKa = 3.88LNLATPEE443 pKa = 4.03DD444 pKa = 3.92QPLGVTLRR452 pKa = 11.84GEE454 pKa = 4.61DD455 pKa = 3.57GDD457 pKa = 4.43GVIVSYY463 pKa = 11.06AVTVPPTRR471 pKa = 11.84GRR473 pKa = 11.84LTGSAPNLTYY483 pKa = 10.32TPQPDD488 pKa = 3.82YY489 pKa = 11.26FGDD492 pKa = 3.54DD493 pKa = 3.13SFTYY497 pKa = 10.13RR498 pKa = 11.84VTDD501 pKa = 3.77DD502 pKa = 3.44VGATATGTVRR512 pKa = 11.84ITVSTVNDD520 pKa = 3.97APTAAADD527 pKa = 3.47SFNAVRR533 pKa = 11.84NALTTVAAPGVLANDD548 pKa = 3.73SDD550 pKa = 4.24IEE552 pKa = 4.29SAAATLTVTATTLATAQDD570 pKa = 3.96GVVTLNADD578 pKa = 3.36GGFTYY583 pKa = 9.84TPPPTAGLTTDD594 pKa = 3.59SFQYY598 pKa = 10.04TLNDD602 pKa = 3.63NDD604 pKa = 4.33PVDD607 pKa = 4.14PLSATGEE614 pKa = 4.14VTLILTGAEE623 pKa = 4.1NLPPSATDD631 pKa = 3.43LTLTTAEE638 pKa = 4.98DD639 pKa = 3.88SPLSITLQGADD650 pKa = 3.29SDD652 pKa = 4.69GAVATYY658 pKa = 9.36TVTTPPQHH666 pKa = 6.15GRR668 pKa = 11.84LTGVAPDD675 pKa = 3.66LTYY678 pKa = 10.86TPDD681 pKa = 3.45ADD683 pKa = 4.04YY684 pKa = 10.79FGSDD688 pKa = 3.02SFVYY692 pKa = 10.18EE693 pKa = 4.1VTDD696 pKa = 3.89SIGAIASATVTLTVNAVNDD715 pKa = 4.21APTAAADD722 pKa = 3.72SFNAARR728 pKa = 11.84NALTTVAAPGVLANDD743 pKa = 3.91SDD745 pKa = 4.47IEE747 pKa = 4.36STPAALTVTAVTLATAQGGAVTLNADD773 pKa = 3.52GGFTYY778 pKa = 9.84TPPPTAGLTTDD789 pKa = 3.59SFQYY793 pKa = 10.04TLNDD797 pKa = 3.63NDD799 pKa = 4.33PVDD802 pKa = 4.14PLSATGEE809 pKa = 4.14VTLILTGAEE818 pKa = 4.1NLPPSATDD826 pKa = 3.43LTLTTAEE833 pKa = 4.98DD834 pKa = 3.88SPLSITLQGTDD845 pKa = 2.5SDD847 pKa = 4.62GAVVAYY853 pKa = 8.33TVTTPPQHH861 pKa = 5.75GQLAGVAPNLIYY873 pKa = 10.54TPAADD878 pKa = 3.84YY879 pKa = 10.45FGSDD883 pKa = 3.03SFVYY887 pKa = 10.18EE888 pKa = 4.1VTDD891 pKa = 3.89SIGAIASATVTLTINAVNDD910 pKa = 3.84APTAAADD917 pKa = 3.72SFNAARR923 pKa = 11.84NTLTTVAAPGVLANDD938 pKa = 3.76SDD940 pKa = 4.25IEE942 pKa = 4.4SVPAEE947 pKa = 4.21LTVTAATLATAQGGAVTLNADD968 pKa = 3.52GGFTYY973 pKa = 9.81TPPVTTGLSTDD984 pKa = 3.23SFQYY988 pKa = 10.07TLNDD992 pKa = 3.63NDD994 pKa = 4.33PVDD997 pKa = 4.14PLSATGEE1004 pKa = 4.14VTLILTGAEE1013 pKa = 4.1NLPPSATDD1021 pKa = 3.43LTLTTAEE1028 pKa = 4.98DD1029 pKa = 3.88SPLSITLQGADD1040 pKa = 2.91SDD1042 pKa = 4.64GAVVAYY1048 pKa = 8.33TVTTPPQHH1056 pKa = 5.75GQLAGVAPNLTYY1068 pKa = 10.53TPAADD1073 pKa = 3.75YY1074 pKa = 10.39FGSDD1078 pKa = 2.98SFIYY1082 pKa = 10.4AVTDD1086 pKa = 3.47NLGATASATVALTISAVNDD1105 pKa = 3.27APTAVDD1111 pKa = 5.37DD1112 pKa = 5.77DD1113 pKa = 4.32FTAQGNIITLIAAPGVLANDD1133 pKa = 3.98SDD1135 pKa = 4.54VEE1137 pKa = 4.39SADD1140 pKa = 3.78ADD1142 pKa = 3.8LTATAEE1148 pKa = 4.52TIATDD1153 pKa = 3.13QGGSVTINTNGSLLYY1168 pKa = 9.59TPPPAAGVTADD1179 pKa = 3.6SFHH1182 pKa = 6.2YY1183 pKa = 9.41TLNDD1187 pKa = 3.39NDD1189 pKa = 4.26PVDD1192 pKa = 4.08PRR1194 pKa = 11.84TSTGTVTIALTTYY1207 pKa = 10.25TEE1209 pKa = 4.24GLSVQGLPAASGDD1222 pKa = 3.82SATARR1227 pKa = 11.84LYY1229 pKa = 11.27LSGTAEE1235 pKa = 4.69SMLSYY1240 pKa = 10.17QWSADD1245 pKa = 3.48GSWVIDD1251 pKa = 3.4SGQGSEE1257 pKa = 4.91VITLTAPAAAGAANVSVVVEE1277 pKa = 4.25DD1278 pKa = 4.45DD1279 pKa = 3.38SGNRR1283 pKa = 11.84AVASLPLSKK1292 pKa = 10.6AGEE1295 pKa = 4.1TAPVLEE1301 pKa = 4.27SLYY1304 pKa = 11.13LEE1306 pKa = 5.09LPPASAPVEE1315 pKa = 4.21LAVTAYY1321 pKa = 10.65DD1322 pKa = 3.96PDD1324 pKa = 4.27GLALDD1329 pKa = 4.36YY1330 pKa = 9.53TWKK1333 pKa = 10.74SAGATVASGATAQWQPPLPGRR1354 pKa = 11.84YY1355 pKa = 9.44QIDD1358 pKa = 3.46IEE1360 pKa = 4.34VDD1362 pKa = 3.18NSSLVAGGSLQRR1374 pKa = 11.84HH1375 pKa = 4.85YY1376 pKa = 10.23TGGAPWAFFRR1386 pKa = 11.84GSRR1389 pKa = 11.84QGTGVRR1395 pKa = 11.84IPADD1399 pKa = 3.37TSSNSGALKK1408 pKa = 8.28WQRR1411 pKa = 11.84PFTMANCAVADD1422 pKa = 3.81NFVPGVVQGEE1432 pKa = 4.4DD1433 pKa = 3.06GTLYY1437 pKa = 10.73AGSSTEE1443 pKa = 3.77GKK1445 pKa = 10.35LYY1447 pKa = 10.89AFNPDD1452 pKa = 3.68DD1453 pKa = 5.89GVIAWSFTTVGTRR1466 pKa = 11.84IDD1468 pKa = 3.88ATPVIAEE1475 pKa = 4.59DD1476 pKa = 3.7GSIYY1480 pKa = 10.52VVEE1483 pKa = 4.4SGGTVYY1489 pKa = 10.83AVNADD1494 pKa = 3.79GSEE1497 pKa = 3.86KK1498 pKa = 9.64WSYY1501 pKa = 11.39AIGAPVSGSPAIGADD1516 pKa = 3.16GTVYY1520 pKa = 10.84VGTDD1524 pKa = 2.88NGGASVLLALNPADD1538 pKa = 5.72GLPKK1542 pKa = 9.9WSAPFALAAEE1552 pKa = 4.62TRR1554 pKa = 11.84SSVNIGADD1562 pKa = 3.3GTVYY1566 pKa = 10.88ARR1568 pKa = 11.84DD1569 pKa = 3.37FAGNLFAINPADD1581 pKa = 3.68GTQRR1585 pKa = 11.84WQITLGGGSGSTPVIAADD1603 pKa = 3.37GTLYY1607 pKa = 10.62FGSFSPPRR1615 pKa = 11.84LYY1617 pKa = 10.95AVGPDD1622 pKa = 4.75GSPKK1626 pKa = 9.62WDD1628 pKa = 3.13TAMGNPSLAGIGATPSVGADD1648 pKa = 3.01GTVYY1652 pKa = 10.71VGVWEE1657 pKa = 4.64AGLNGSVYY1665 pKa = 10.68AIDD1668 pKa = 3.85PVGGTVVWRR1677 pKa = 11.84RR1678 pKa = 11.84QLDD1681 pKa = 3.73SFVQSAATAVGADD1694 pKa = 3.06GRR1696 pKa = 11.84VYY1698 pKa = 10.22VVSARR1703 pKa = 11.84GVFYY1707 pKa = 10.92ALDD1710 pKa = 4.48PDD1712 pKa = 3.88NGAVDD1717 pKa = 3.38WSFDD1721 pKa = 2.8IGAATNEE1728 pKa = 4.21EE1729 pKa = 4.74SPSPLAIGTDD1739 pKa = 3.54GSIYY1743 pKa = 10.57VYY1745 pKa = 10.26TCDD1748 pKa = 3.12GVLRR1752 pKa = 11.84AFQQ1755 pKa = 3.44
Molecular weight: 178.6 kDa
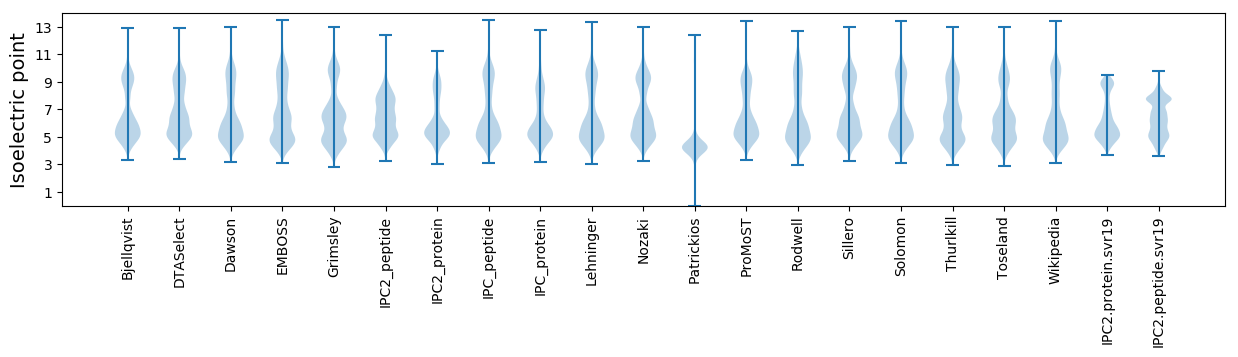
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A545U3L0|A0A545U3L0_9GAMM Uncharacterized protein OS=Exilibacterium tricleocarpae OX=2591008 GN=FKG94_05190 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.25RR14 pKa = 11.84NHH16 pKa = 5.6GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.46NGRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.31QLAAA44 pKa = 4.14
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.25RR14 pKa = 11.84NHH16 pKa = 5.6GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.46NGRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.31QLAAA44 pKa = 4.14
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2009629 |
18 |
12098 |
361.7 |
39.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.475 ± 0.053 | 1.036 ± 0.012 |
6.002 ± 0.034 | 5.701 ± 0.029 |
3.855 ± 0.022 | 7.86 ± 0.031 |
2.148 ± 0.017 | 5.216 ± 0.029 |
3.544 ± 0.036 | 10.442 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.025 ± 0.019 | 3.551 ± 0.026 |
4.59 ± 0.025 | 4.198 ± 0.025 |
6.349 ± 0.028 | 5.93 ± 0.028 |
5.557 ± 0.034 | 7.169 ± 0.027 |
1.329 ± 0.013 | 3.025 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
