
Capybara microvirus Cap1_SP_119
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
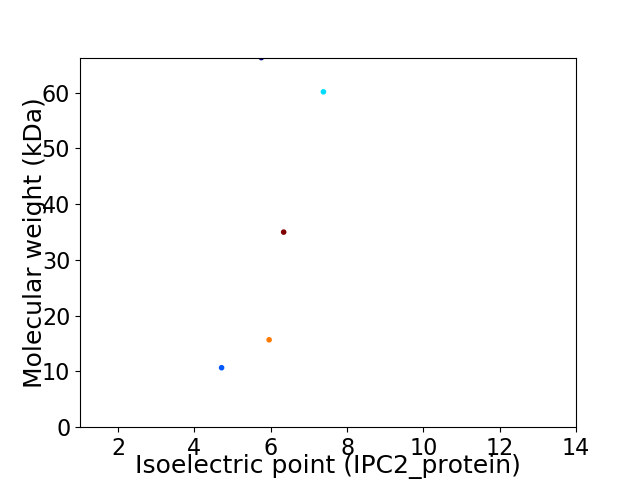
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W471|A0A4P8W471_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_119 OX=2584811 PE=4 SV=1
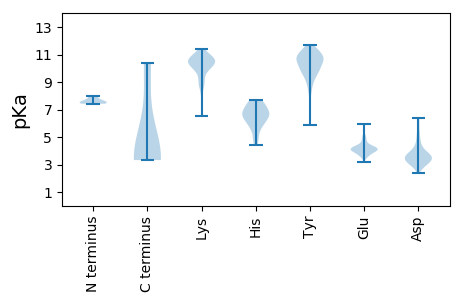
MM1 pKa = 7.64KK2 pKa = 9.77PVNYY6 pKa = 9.86FNRR9 pKa = 11.84DD10 pKa = 3.28CITPKK15 pKa = 10.55NEE17 pKa = 3.98EE18 pKa = 5.07LICQSKK24 pKa = 10.62DD25 pKa = 2.74GKK27 pKa = 9.12TFFAPFPSVVAQAKK41 pKa = 10.03DD42 pKa = 3.51YY43 pKa = 11.27SLEE46 pKa = 4.04SLLAANIDD54 pKa = 3.6PSKK57 pKa = 11.04LVIKK61 pKa = 9.83VKK63 pKa = 10.84KK64 pKa = 10.48EE65 pKa = 3.6GVTDD69 pKa = 4.32FDD71 pKa = 3.97EE72 pKa = 4.28EE73 pKa = 4.84TIVVRR78 pKa = 11.84LNSIVEE84 pKa = 4.17NLDD87 pKa = 3.31EE88 pKa = 4.71SNQPNEE94 pKa = 4.11SS95 pKa = 3.32
MM1 pKa = 7.64KK2 pKa = 9.77PVNYY6 pKa = 9.86FNRR9 pKa = 11.84DD10 pKa = 3.28CITPKK15 pKa = 10.55NEE17 pKa = 3.98EE18 pKa = 5.07LICQSKK24 pKa = 10.62DD25 pKa = 2.74GKK27 pKa = 9.12TFFAPFPSVVAQAKK41 pKa = 10.03DD42 pKa = 3.51YY43 pKa = 11.27SLEE46 pKa = 4.04SLLAANIDD54 pKa = 3.6PSKK57 pKa = 11.04LVIKK61 pKa = 9.83VKK63 pKa = 10.84KK64 pKa = 10.48EE65 pKa = 3.6GVTDD69 pKa = 4.32FDD71 pKa = 3.97EE72 pKa = 4.28EE73 pKa = 4.84TIVVRR78 pKa = 11.84LNSIVEE84 pKa = 4.17NLDD87 pKa = 3.31EE88 pKa = 4.71SNQPNEE94 pKa = 4.11SS95 pKa = 3.32
Molecular weight: 10.67 kDa
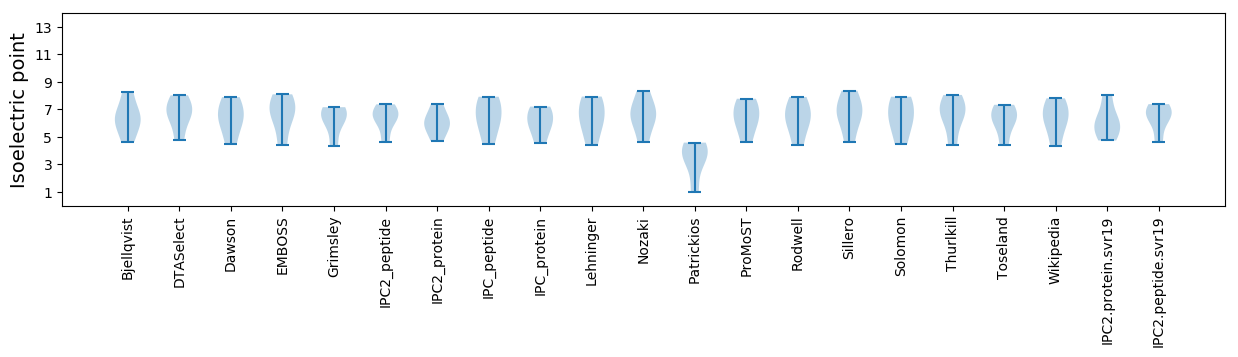
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7V1|A0A4P8W7V1_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_119 OX=2584811 PE=3 SV=1
MM1 pKa = 7.98KK2 pKa = 9.94IRR4 pKa = 11.84NKK6 pKa = 9.43STFYY10 pKa = 10.78SDD12 pKa = 3.42YY13 pKa = 10.97SKK15 pKa = 8.89THH17 pKa = 6.29FDD19 pKa = 3.08IDD21 pKa = 2.92TDD23 pKa = 3.61IFVRR27 pKa = 11.84SCSSVYY33 pKa = 10.34SYY35 pKa = 10.94SVRR38 pKa = 11.84NYY40 pKa = 9.66YY41 pKa = 9.79EE42 pKa = 4.03YY43 pKa = 11.27KK44 pKa = 10.59SVIDD48 pKa = 4.06SGGQCFFYY56 pKa = 10.63TLTHH60 pKa = 6.56NDD62 pKa = 2.97SSIVRR67 pKa = 11.84LYY69 pKa = 10.92GRR71 pKa = 11.84SFPNYY76 pKa = 8.29EE77 pKa = 3.88GLRR80 pKa = 11.84NLVNGAFKK88 pKa = 10.7KK89 pKa = 10.48RR90 pKa = 11.84IEE92 pKa = 4.15KK93 pKa = 8.86LTNCRR98 pKa = 11.84FTYY101 pKa = 10.09FAVTEE106 pKa = 4.3FGEE109 pKa = 4.67GKK111 pKa = 10.45GYY113 pKa = 10.57RR114 pKa = 11.84GLDD117 pKa = 3.62NNPHH121 pKa = 4.44YY122 pKa = 10.87HH123 pKa = 6.03ILFYY127 pKa = 10.31LHH129 pKa = 7.39PCTEE133 pKa = 4.1KK134 pKa = 10.83GSYY137 pKa = 10.22VPISSNDD144 pKa = 2.84FCEE147 pKa = 3.81LVRR150 pKa = 11.84YY151 pKa = 8.07YY152 pKa = 9.92WQGDD156 pKa = 3.32KK157 pKa = 11.13YY158 pKa = 10.47HH159 pKa = 6.73YY160 pKa = 10.15KK161 pKa = 10.65DD162 pKa = 3.58SLHH165 pKa = 7.26ILEE168 pKa = 5.13PKK170 pKa = 10.38DD171 pKa = 3.75YY172 pKa = 9.8IYY174 pKa = 11.1GIASPSNINNGEE186 pKa = 4.01VTNFSAIRR194 pKa = 11.84YY195 pKa = 5.91CCKK198 pKa = 10.65YY199 pKa = 8.18ITKK202 pKa = 10.32DD203 pKa = 2.38SYY205 pKa = 11.22YY206 pKa = 10.48QVFRR210 pKa = 11.84KK211 pKa = 9.97NLLKK215 pKa = 10.47DD216 pKa = 3.51IYY218 pKa = 10.9YY219 pKa = 10.57SIVKK223 pKa = 9.97QLTLCDD229 pKa = 5.33DD230 pKa = 3.98IDD232 pKa = 3.77IQTDD236 pKa = 3.52PNYY239 pKa = 9.4VTSRR243 pKa = 11.84LISFCGSLNTAFDD256 pKa = 5.4FINEE260 pKa = 4.1QQKK263 pKa = 10.71KK264 pKa = 9.07IYY266 pKa = 9.49CSLYY270 pKa = 9.44SYY272 pKa = 10.5IHH274 pKa = 6.64LVHH277 pKa = 6.78CSKK280 pKa = 11.16KK281 pKa = 10.46LGSLGLEE288 pKa = 4.27YY289 pKa = 9.6IDD291 pKa = 4.8YY292 pKa = 10.89DD293 pKa = 4.15NNSLTIPFPDD303 pKa = 3.11SKK305 pKa = 11.37GSLRR309 pKa = 11.84QCSLPLYY316 pKa = 9.46YY317 pKa = 10.05YY318 pKa = 10.67RR319 pKa = 11.84KK320 pKa = 8.78VCYY323 pKa = 10.02DD324 pKa = 2.67ITKK327 pKa = 9.39NYY329 pKa = 9.27RR330 pKa = 11.84GNVHH334 pKa = 6.6YY335 pKa = 10.41YY336 pKa = 10.83LNDD339 pKa = 3.35YY340 pKa = 10.26GIKK343 pKa = 10.11CKK345 pKa = 10.93SNLFPKK351 pKa = 10.22DD352 pKa = 3.35LKK354 pKa = 11.08KK355 pKa = 10.83QFDD358 pKa = 3.59ICKK361 pKa = 8.58STISSYY367 pKa = 11.17FDD369 pKa = 5.2DD370 pKa = 6.36DD371 pKa = 5.83LLTDD375 pKa = 3.83YY376 pKa = 11.64NNLHH380 pKa = 6.61INSSFTYY387 pKa = 10.56QDD389 pKa = 3.06IKK391 pKa = 11.31DD392 pKa = 4.1LYY394 pKa = 11.24HH395 pKa = 7.45NDD397 pKa = 3.67EE398 pKa = 4.59LLNAYY403 pKa = 9.68VLFNFVYY410 pKa = 10.37RR411 pKa = 11.84EE412 pKa = 3.55RR413 pKa = 11.84FYY415 pKa = 11.37DD416 pKa = 3.34RR417 pKa = 11.84RR418 pKa = 11.84HH419 pKa = 5.77ISICPEE425 pKa = 3.21FDD427 pKa = 3.44YY428 pKa = 11.51EE429 pKa = 4.33HH430 pKa = 6.93FLVPSLVEE438 pKa = 3.97TPFTHH443 pKa = 7.54IEE445 pKa = 4.1PNLSKK450 pKa = 11.27YY451 pKa = 9.96EE452 pKa = 4.14SYY454 pKa = 11.38EE455 pKa = 3.63NNPYY459 pKa = 10.11FKK461 pKa = 10.42PYY463 pKa = 8.84MPKK466 pKa = 10.45FSLLDD471 pKa = 3.4EE472 pKa = 5.0LFTFFKK478 pKa = 9.94KK479 pKa = 8.61TAQEE483 pKa = 4.09YY484 pKa = 10.16KK485 pKa = 9.01IKK487 pKa = 10.21EE488 pKa = 4.03YY489 pKa = 10.58EE490 pKa = 3.65RR491 pKa = 11.84RR492 pKa = 11.84KK493 pKa = 8.88YY494 pKa = 10.47LKK496 pKa = 9.98RR497 pKa = 11.84VHH499 pKa = 5.66STQNII504 pKa = 3.46
MM1 pKa = 7.98KK2 pKa = 9.94IRR4 pKa = 11.84NKK6 pKa = 9.43STFYY10 pKa = 10.78SDD12 pKa = 3.42YY13 pKa = 10.97SKK15 pKa = 8.89THH17 pKa = 6.29FDD19 pKa = 3.08IDD21 pKa = 2.92TDD23 pKa = 3.61IFVRR27 pKa = 11.84SCSSVYY33 pKa = 10.34SYY35 pKa = 10.94SVRR38 pKa = 11.84NYY40 pKa = 9.66YY41 pKa = 9.79EE42 pKa = 4.03YY43 pKa = 11.27KK44 pKa = 10.59SVIDD48 pKa = 4.06SGGQCFFYY56 pKa = 10.63TLTHH60 pKa = 6.56NDD62 pKa = 2.97SSIVRR67 pKa = 11.84LYY69 pKa = 10.92GRR71 pKa = 11.84SFPNYY76 pKa = 8.29EE77 pKa = 3.88GLRR80 pKa = 11.84NLVNGAFKK88 pKa = 10.7KK89 pKa = 10.48RR90 pKa = 11.84IEE92 pKa = 4.15KK93 pKa = 8.86LTNCRR98 pKa = 11.84FTYY101 pKa = 10.09FAVTEE106 pKa = 4.3FGEE109 pKa = 4.67GKK111 pKa = 10.45GYY113 pKa = 10.57RR114 pKa = 11.84GLDD117 pKa = 3.62NNPHH121 pKa = 4.44YY122 pKa = 10.87HH123 pKa = 6.03ILFYY127 pKa = 10.31LHH129 pKa = 7.39PCTEE133 pKa = 4.1KK134 pKa = 10.83GSYY137 pKa = 10.22VPISSNDD144 pKa = 2.84FCEE147 pKa = 3.81LVRR150 pKa = 11.84YY151 pKa = 8.07YY152 pKa = 9.92WQGDD156 pKa = 3.32KK157 pKa = 11.13YY158 pKa = 10.47HH159 pKa = 6.73YY160 pKa = 10.15KK161 pKa = 10.65DD162 pKa = 3.58SLHH165 pKa = 7.26ILEE168 pKa = 5.13PKK170 pKa = 10.38DD171 pKa = 3.75YY172 pKa = 9.8IYY174 pKa = 11.1GIASPSNINNGEE186 pKa = 4.01VTNFSAIRR194 pKa = 11.84YY195 pKa = 5.91CCKK198 pKa = 10.65YY199 pKa = 8.18ITKK202 pKa = 10.32DD203 pKa = 2.38SYY205 pKa = 11.22YY206 pKa = 10.48QVFRR210 pKa = 11.84KK211 pKa = 9.97NLLKK215 pKa = 10.47DD216 pKa = 3.51IYY218 pKa = 10.9YY219 pKa = 10.57SIVKK223 pKa = 9.97QLTLCDD229 pKa = 5.33DD230 pKa = 3.98IDD232 pKa = 3.77IQTDD236 pKa = 3.52PNYY239 pKa = 9.4VTSRR243 pKa = 11.84LISFCGSLNTAFDD256 pKa = 5.4FINEE260 pKa = 4.1QQKK263 pKa = 10.71KK264 pKa = 9.07IYY266 pKa = 9.49CSLYY270 pKa = 9.44SYY272 pKa = 10.5IHH274 pKa = 6.64LVHH277 pKa = 6.78CSKK280 pKa = 11.16KK281 pKa = 10.46LGSLGLEE288 pKa = 4.27YY289 pKa = 9.6IDD291 pKa = 4.8YY292 pKa = 10.89DD293 pKa = 4.15NNSLTIPFPDD303 pKa = 3.11SKK305 pKa = 11.37GSLRR309 pKa = 11.84QCSLPLYY316 pKa = 9.46YY317 pKa = 10.05YY318 pKa = 10.67RR319 pKa = 11.84KK320 pKa = 8.78VCYY323 pKa = 10.02DD324 pKa = 2.67ITKK327 pKa = 9.39NYY329 pKa = 9.27RR330 pKa = 11.84GNVHH334 pKa = 6.6YY335 pKa = 10.41YY336 pKa = 10.83LNDD339 pKa = 3.35YY340 pKa = 10.26GIKK343 pKa = 10.11CKK345 pKa = 10.93SNLFPKK351 pKa = 10.22DD352 pKa = 3.35LKK354 pKa = 11.08KK355 pKa = 10.83QFDD358 pKa = 3.59ICKK361 pKa = 8.58STISSYY367 pKa = 11.17FDD369 pKa = 5.2DD370 pKa = 6.36DD371 pKa = 5.83LLTDD375 pKa = 3.83YY376 pKa = 11.64NNLHH380 pKa = 6.61INSSFTYY387 pKa = 10.56QDD389 pKa = 3.06IKK391 pKa = 11.31DD392 pKa = 4.1LYY394 pKa = 11.24HH395 pKa = 7.45NDD397 pKa = 3.67EE398 pKa = 4.59LLNAYY403 pKa = 9.68VLFNFVYY410 pKa = 10.37RR411 pKa = 11.84EE412 pKa = 3.55RR413 pKa = 11.84FYY415 pKa = 11.37DD416 pKa = 3.34RR417 pKa = 11.84RR418 pKa = 11.84HH419 pKa = 5.77ISICPEE425 pKa = 3.21FDD427 pKa = 3.44YY428 pKa = 11.51EE429 pKa = 4.33HH430 pKa = 6.93FLVPSLVEE438 pKa = 3.97TPFTHH443 pKa = 7.54IEE445 pKa = 4.1PNLSKK450 pKa = 11.27YY451 pKa = 9.96EE452 pKa = 4.14SYY454 pKa = 11.38EE455 pKa = 3.63NNPYY459 pKa = 10.11FKK461 pKa = 10.42PYY463 pKa = 8.84MPKK466 pKa = 10.45FSLLDD471 pKa = 3.4EE472 pKa = 5.0LFTFFKK478 pKa = 9.94KK479 pKa = 8.61TAQEE483 pKa = 4.09YY484 pKa = 10.16KK485 pKa = 9.01IKK487 pKa = 10.21EE488 pKa = 4.03YY489 pKa = 10.58EE490 pKa = 3.65RR491 pKa = 11.84RR492 pKa = 11.84KK493 pKa = 8.88YY494 pKa = 10.47LKK496 pKa = 9.98RR497 pKa = 11.84VHH499 pKa = 5.66STQNII504 pKa = 3.46
Molecular weight: 60.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1633 |
95 |
583 |
326.6 |
37.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.735 ± 0.885 | 1.715 ± 0.642 |
5.94 ± 0.587 | 5.266 ± 0.647 |
6.185 ± 0.625 | 4.838 ± 0.816 |
1.776 ± 0.538 | 6.491 ± 0.252 |
6.552 ± 0.678 | 8.757 ± 0.572 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.653 ± 0.571 | 7.655 ± 0.307 |
3.429 ± 0.411 | 3.184 ± 0.587 |
4.287 ± 0.368 | 10.533 ± 0.882 |
4.654 ± 0.354 | 6.062 ± 0.711 |
0.367 ± 0.114 | 6.92 ± 1.51 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
