
Sulfitobacter donghicola DSW-25 = KCTC 12864 = JCM 14565
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Sulfitobacter; Sulfitobacter donghicola
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
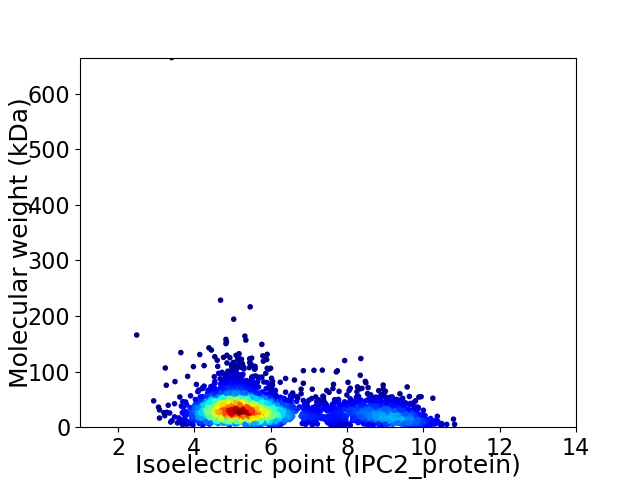
Virtual 2D-PAGE plot for 3416 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A073IGL0|A0A073IGL0_9RHOB Glycine/betaine ABC transporter OS=Sulfitobacter donghicola DSW-25 = KCTC 12864 = JCM 14565 OX=1300350 GN=DSW25_13230 PE=3 SV=1
MM1 pKa = 8.08IEE3 pKa = 4.09VEE5 pKa = 4.27LLQAIEE11 pKa = 4.3SGQNVTSGHH20 pKa = 6.09FGGNVIAGKK29 pKa = 8.61NTDD32 pKa = 4.02DD33 pKa = 6.16GIPTEE38 pKa = 4.32QNSLAHH44 pKa = 6.16QEE46 pKa = 3.81LDD48 pKa = 3.27IEE50 pKa = 4.62VARR53 pKa = 11.84YY54 pKa = 7.69PAGEE58 pKa = 4.53PDD60 pKa = 4.81LMYY63 pKa = 10.84KK64 pKa = 10.5DD65 pKa = 4.01GMVINGALPDD75 pKa = 3.79HH76 pKa = 7.16LINFLMAARR85 pKa = 11.84ASGQSVVLVTPTHH98 pKa = 6.2EE99 pKa = 5.18GYY101 pKa = 10.5HH102 pKa = 5.9GADD105 pKa = 2.97ILTEE109 pKa = 4.15FTSLVLGQFSDD120 pKa = 4.14VIHH123 pKa = 6.63AFEE126 pKa = 5.61IGNEE130 pKa = 4.15YY131 pKa = 9.98WNHH134 pKa = 4.16QTEE137 pKa = 4.19TSYY140 pKa = 12.0GEE142 pKa = 4.07VANDD146 pKa = 3.22SVLAISSQLTGQYY159 pKa = 10.48GDD161 pKa = 3.21IPIWVQMGDD170 pKa = 3.04AGGQASEE177 pKa = 4.34FAKK180 pKa = 10.54DD181 pKa = 3.38APLSEE186 pKa = 4.91GIGWLWRR193 pKa = 11.84NIGANNLILDD203 pKa = 3.79QLTPEE208 pKa = 4.08ARR210 pKa = 11.84ATIDD214 pKa = 3.34GVVEE218 pKa = 4.12HH219 pKa = 6.67YY220 pKa = 10.55YY221 pKa = 10.65FRR223 pKa = 11.84EE224 pKa = 3.75DD225 pKa = 3.03HH226 pKa = 6.51QYY228 pKa = 11.33LGFYY232 pKa = 10.11NDD234 pKa = 3.32QNIVMDD240 pKa = 3.94HH241 pKa = 6.12EE242 pKa = 4.94VWQSAFGRR250 pKa = 11.84NLTLNITEE258 pKa = 4.16WNIRR262 pKa = 11.84TTNLDD267 pKa = 3.19QLGIRR272 pKa = 11.84AASTLIAQFSFMMDD286 pKa = 3.28MEE288 pKa = 4.27VDD290 pKa = 3.57EE291 pKa = 5.74AYY293 pKa = 10.35VWPPMHH299 pKa = 6.23NTSTDD304 pKa = 3.33LAGSSNVLLDD314 pKa = 3.76PEE316 pKa = 4.41TGIVINSVGGATFDD330 pKa = 3.95LMSSSLVGLQYY341 pKa = 10.55RR342 pKa = 11.84PSATTSDD349 pKa = 3.11SSLLHH354 pKa = 6.17NYY356 pKa = 9.26VYY358 pKa = 10.72SGEE361 pKa = 4.35DD362 pKa = 2.94KK363 pKa = 10.72VVVYY367 pKa = 7.6VTSRR371 pKa = 11.84SDD373 pKa = 3.21EE374 pKa = 4.3TEE376 pKa = 3.82SVSFNLGNFFPGASLISATQIGYY399 pKa = 10.26DD400 pKa = 3.44KK401 pKa = 10.37TSSDD405 pKa = 3.43GRR407 pKa = 11.84HH408 pKa = 5.31YY409 pKa = 11.13DD410 pKa = 3.36YY411 pKa = 11.26VQRR414 pKa = 11.84EE415 pKa = 4.35FVDD418 pKa = 3.97SEE420 pKa = 4.01AVAIDD425 pKa = 3.52GEE427 pKa = 4.4LYY429 pKa = 8.87YY430 pKa = 10.88TNEE433 pKa = 3.79HH434 pKa = 6.79DD435 pKa = 3.95VRR437 pKa = 11.84ATITEE442 pKa = 4.32HH443 pKa = 6.9DD444 pKa = 3.96TSQSTNGGNFTFTLLPYY461 pKa = 10.21EE462 pKa = 4.55VIEE465 pKa = 4.14LTYY468 pKa = 9.85EE469 pKa = 3.63IPNFEE474 pKa = 4.42RR475 pKa = 11.84TDD477 pKa = 3.44GTNGKK482 pKa = 9.86DD483 pKa = 3.2IISSEE488 pKa = 4.04DD489 pKa = 3.32SKK491 pKa = 11.92NDD493 pKa = 3.5LVFSLAGNDD502 pKa = 3.7NIQAGSGDD510 pKa = 3.63DD511 pKa = 3.79SIYY514 pKa = 11.17AGDD517 pKa = 3.87GDD519 pKa = 4.87DD520 pKa = 5.65YY521 pKa = 11.62IDD523 pKa = 5.23AGLGDD528 pKa = 4.66DD529 pKa = 4.62LVSGGAGNDD538 pKa = 3.52IIRR541 pKa = 11.84GRR543 pKa = 11.84GNNDD547 pKa = 3.33IISGGSGNDD556 pKa = 4.37DD557 pKa = 3.47IDD559 pKa = 4.99GGWGDD564 pKa = 4.66DD565 pKa = 3.97KK566 pKa = 11.73LDD568 pKa = 4.06GGSGSDD574 pKa = 3.5ILNGGWGRR582 pKa = 11.84DD583 pKa = 3.64TITLDD588 pKa = 3.11SSLDD592 pKa = 3.4IAIGGGDD599 pKa = 4.26DD600 pKa = 6.54DD601 pKa = 6.27LFQLDD606 pKa = 3.9GLQKK610 pKa = 10.87YY611 pKa = 8.21GAGWGGINASNQYY624 pKa = 10.51QDD626 pKa = 2.91GTMVFLPVANYY637 pKa = 10.23NLYY640 pKa = 10.64SSVIIGGAGYY650 pKa = 8.22DD651 pKa = 3.89TIEE654 pKa = 4.98LSASDD659 pKa = 4.66DD660 pKa = 4.89AIFLHH665 pKa = 6.56NGFSSLPSSLNVSEE679 pKa = 4.58YY680 pKa = 11.26VSDD683 pKa = 4.07LPSGLMISGIEE694 pKa = 4.06EE695 pKa = 3.77FRR697 pKa = 11.84AGAGNDD703 pKa = 3.8FVDD706 pKa = 3.97LTSDD710 pKa = 3.57VFHH713 pKa = 6.89LQGQSIVIHH722 pKa = 6.69GDD724 pKa = 3.11YY725 pKa = 10.93GHH727 pKa = 7.0DD728 pKa = 3.95VVWGSAANEE737 pKa = 4.34TIFGGEE743 pKa = 4.19GNDD746 pKa = 3.7TLFGGAGNDD755 pKa = 3.82EE756 pKa = 4.22ISGGTGADD764 pKa = 3.08VFEE767 pKa = 4.52FTSTSIDD774 pKa = 3.24TSLTDD779 pKa = 3.61FDD781 pKa = 4.12VRR783 pKa = 11.84EE784 pKa = 4.09GDD786 pKa = 3.39EE787 pKa = 3.76LRR789 pKa = 11.84FYY791 pKa = 10.13NTEE794 pKa = 3.76NVEE797 pKa = 3.95FDD799 pKa = 3.59AGSVALTPGGINISYY814 pKa = 10.07RR815 pKa = 11.84NTVSEE820 pKa = 4.27TEE822 pKa = 3.99HH823 pKa = 7.37YY824 pKa = 10.72IFISLVTHH832 pKa = 5.1STEE835 pKa = 4.06FAATLPEE842 pKa = 4.11ILNALEE848 pKa = 4.16IVV850 pKa = 3.55
MM1 pKa = 8.08IEE3 pKa = 4.09VEE5 pKa = 4.27LLQAIEE11 pKa = 4.3SGQNVTSGHH20 pKa = 6.09FGGNVIAGKK29 pKa = 8.61NTDD32 pKa = 4.02DD33 pKa = 6.16GIPTEE38 pKa = 4.32QNSLAHH44 pKa = 6.16QEE46 pKa = 3.81LDD48 pKa = 3.27IEE50 pKa = 4.62VARR53 pKa = 11.84YY54 pKa = 7.69PAGEE58 pKa = 4.53PDD60 pKa = 4.81LMYY63 pKa = 10.84KK64 pKa = 10.5DD65 pKa = 4.01GMVINGALPDD75 pKa = 3.79HH76 pKa = 7.16LINFLMAARR85 pKa = 11.84ASGQSVVLVTPTHH98 pKa = 6.2EE99 pKa = 5.18GYY101 pKa = 10.5HH102 pKa = 5.9GADD105 pKa = 2.97ILTEE109 pKa = 4.15FTSLVLGQFSDD120 pKa = 4.14VIHH123 pKa = 6.63AFEE126 pKa = 5.61IGNEE130 pKa = 4.15YY131 pKa = 9.98WNHH134 pKa = 4.16QTEE137 pKa = 4.19TSYY140 pKa = 12.0GEE142 pKa = 4.07VANDD146 pKa = 3.22SVLAISSQLTGQYY159 pKa = 10.48GDD161 pKa = 3.21IPIWVQMGDD170 pKa = 3.04AGGQASEE177 pKa = 4.34FAKK180 pKa = 10.54DD181 pKa = 3.38APLSEE186 pKa = 4.91GIGWLWRR193 pKa = 11.84NIGANNLILDD203 pKa = 3.79QLTPEE208 pKa = 4.08ARR210 pKa = 11.84ATIDD214 pKa = 3.34GVVEE218 pKa = 4.12HH219 pKa = 6.67YY220 pKa = 10.55YY221 pKa = 10.65FRR223 pKa = 11.84EE224 pKa = 3.75DD225 pKa = 3.03HH226 pKa = 6.51QYY228 pKa = 11.33LGFYY232 pKa = 10.11NDD234 pKa = 3.32QNIVMDD240 pKa = 3.94HH241 pKa = 6.12EE242 pKa = 4.94VWQSAFGRR250 pKa = 11.84NLTLNITEE258 pKa = 4.16WNIRR262 pKa = 11.84TTNLDD267 pKa = 3.19QLGIRR272 pKa = 11.84AASTLIAQFSFMMDD286 pKa = 3.28MEE288 pKa = 4.27VDD290 pKa = 3.57EE291 pKa = 5.74AYY293 pKa = 10.35VWPPMHH299 pKa = 6.23NTSTDD304 pKa = 3.33LAGSSNVLLDD314 pKa = 3.76PEE316 pKa = 4.41TGIVINSVGGATFDD330 pKa = 3.95LMSSSLVGLQYY341 pKa = 10.55RR342 pKa = 11.84PSATTSDD349 pKa = 3.11SSLLHH354 pKa = 6.17NYY356 pKa = 9.26VYY358 pKa = 10.72SGEE361 pKa = 4.35DD362 pKa = 2.94KK363 pKa = 10.72VVVYY367 pKa = 7.6VTSRR371 pKa = 11.84SDD373 pKa = 3.21EE374 pKa = 4.3TEE376 pKa = 3.82SVSFNLGNFFPGASLISATQIGYY399 pKa = 10.26DD400 pKa = 3.44KK401 pKa = 10.37TSSDD405 pKa = 3.43GRR407 pKa = 11.84HH408 pKa = 5.31YY409 pKa = 11.13DD410 pKa = 3.36YY411 pKa = 11.26VQRR414 pKa = 11.84EE415 pKa = 4.35FVDD418 pKa = 3.97SEE420 pKa = 4.01AVAIDD425 pKa = 3.52GEE427 pKa = 4.4LYY429 pKa = 8.87YY430 pKa = 10.88TNEE433 pKa = 3.79HH434 pKa = 6.79DD435 pKa = 3.95VRR437 pKa = 11.84ATITEE442 pKa = 4.32HH443 pKa = 6.9DD444 pKa = 3.96TSQSTNGGNFTFTLLPYY461 pKa = 10.21EE462 pKa = 4.55VIEE465 pKa = 4.14LTYY468 pKa = 9.85EE469 pKa = 3.63IPNFEE474 pKa = 4.42RR475 pKa = 11.84TDD477 pKa = 3.44GTNGKK482 pKa = 9.86DD483 pKa = 3.2IISSEE488 pKa = 4.04DD489 pKa = 3.32SKK491 pKa = 11.92NDD493 pKa = 3.5LVFSLAGNDD502 pKa = 3.7NIQAGSGDD510 pKa = 3.63DD511 pKa = 3.79SIYY514 pKa = 11.17AGDD517 pKa = 3.87GDD519 pKa = 4.87DD520 pKa = 5.65YY521 pKa = 11.62IDD523 pKa = 5.23AGLGDD528 pKa = 4.66DD529 pKa = 4.62LVSGGAGNDD538 pKa = 3.52IIRR541 pKa = 11.84GRR543 pKa = 11.84GNNDD547 pKa = 3.33IISGGSGNDD556 pKa = 4.37DD557 pKa = 3.47IDD559 pKa = 4.99GGWGDD564 pKa = 4.66DD565 pKa = 3.97KK566 pKa = 11.73LDD568 pKa = 4.06GGSGSDD574 pKa = 3.5ILNGGWGRR582 pKa = 11.84DD583 pKa = 3.64TITLDD588 pKa = 3.11SSLDD592 pKa = 3.4IAIGGGDD599 pKa = 4.26DD600 pKa = 6.54DD601 pKa = 6.27LFQLDD606 pKa = 3.9GLQKK610 pKa = 10.87YY611 pKa = 8.21GAGWGGINASNQYY624 pKa = 10.51QDD626 pKa = 2.91GTMVFLPVANYY637 pKa = 10.23NLYY640 pKa = 10.64SSVIIGGAGYY650 pKa = 8.22DD651 pKa = 3.89TIEE654 pKa = 4.98LSASDD659 pKa = 4.66DD660 pKa = 4.89AIFLHH665 pKa = 6.56NGFSSLPSSLNVSEE679 pKa = 4.58YY680 pKa = 11.26VSDD683 pKa = 4.07LPSGLMISGIEE694 pKa = 4.06EE695 pKa = 3.77FRR697 pKa = 11.84AGAGNDD703 pKa = 3.8FVDD706 pKa = 3.97LTSDD710 pKa = 3.57VFHH713 pKa = 6.89LQGQSIVIHH722 pKa = 6.69GDD724 pKa = 3.11YY725 pKa = 10.93GHH727 pKa = 7.0DD728 pKa = 3.95VVWGSAANEE737 pKa = 4.34TIFGGEE743 pKa = 4.19GNDD746 pKa = 3.7TLFGGAGNDD755 pKa = 3.82EE756 pKa = 4.22ISGGTGADD764 pKa = 3.08VFEE767 pKa = 4.52FTSTSIDD774 pKa = 3.24TSLTDD779 pKa = 3.61FDD781 pKa = 4.12VRR783 pKa = 11.84EE784 pKa = 4.09GDD786 pKa = 3.39EE787 pKa = 3.76LRR789 pKa = 11.84FYY791 pKa = 10.13NTEE794 pKa = 3.76NVEE797 pKa = 3.95FDD799 pKa = 3.59AGSVALTPGGINISYY814 pKa = 10.07RR815 pKa = 11.84NTVSEE820 pKa = 4.27TEE822 pKa = 3.99HH823 pKa = 7.37YY824 pKa = 10.72IFISLVTHH832 pKa = 5.1STEE835 pKa = 4.06FAATLPEE842 pKa = 4.11ILNALEE848 pKa = 4.16IVV850 pKa = 3.55
Molecular weight: 91.46 kDa
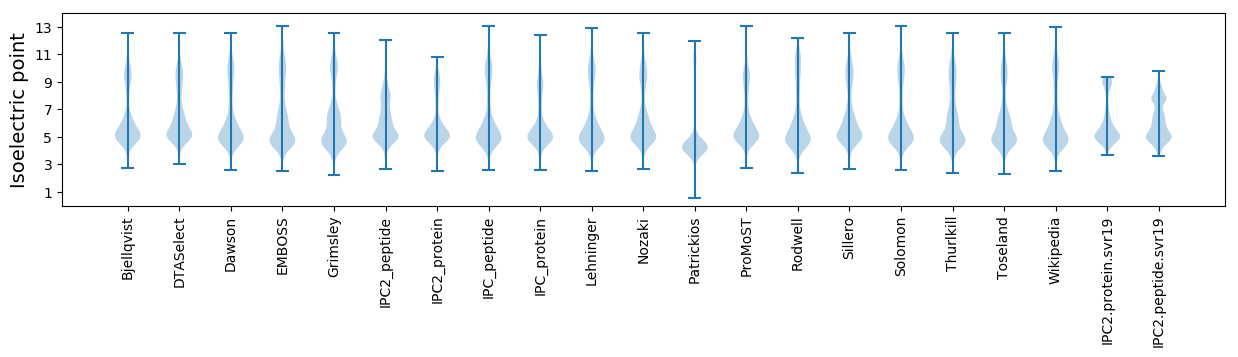
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A073IFB4|A0A073IFB4_9RHOB Radical SAM protein OS=Sulfitobacter donghicola DSW-25 = KCTC 12864 = JCM 14565 OX=1300350 GN=DSW25_15235 PE=4 SV=1
MM1 pKa = 7.42NLTAFANASPVIQIHH16 pKa = 5.32TLLALGAVLLTISIFALRR34 pKa = 11.84KK35 pKa = 9.17GSPLHH40 pKa = 6.93RR41 pKa = 11.84VMGWTWVIMMALVALSSFWINEE63 pKa = 3.57LRR65 pKa = 11.84VLGAFSPIHH74 pKa = 6.2LLSIITLVTLVSSVTAARR92 pKa = 11.84RR93 pKa = 11.84HH94 pKa = 4.2NVKK97 pKa = 9.62RR98 pKa = 11.84HH99 pKa = 4.21KK100 pKa = 10.36RR101 pKa = 11.84QMRR104 pKa = 11.84TLVFGALIVAGGFTFLPGRR123 pKa = 11.84LMHH126 pKa = 6.07TVFLGGG132 pKa = 3.45
MM1 pKa = 7.42NLTAFANASPVIQIHH16 pKa = 5.32TLLALGAVLLTISIFALRR34 pKa = 11.84KK35 pKa = 9.17GSPLHH40 pKa = 6.93RR41 pKa = 11.84VMGWTWVIMMALVALSSFWINEE63 pKa = 3.57LRR65 pKa = 11.84VLGAFSPIHH74 pKa = 6.2LLSIITLVTLVSSVTAARR92 pKa = 11.84RR93 pKa = 11.84HH94 pKa = 4.2NVKK97 pKa = 9.62RR98 pKa = 11.84HH99 pKa = 4.21KK100 pKa = 10.36RR101 pKa = 11.84QMRR104 pKa = 11.84TLVFGALIVAGGFTFLPGRR123 pKa = 11.84LMHH126 pKa = 6.07TVFLGGG132 pKa = 3.45
Molecular weight: 14.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1055224 |
29 |
6491 |
308.9 |
33.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.669 ± 0.049 | 0.884 ± 0.014 |
6.096 ± 0.041 | 6.075 ± 0.04 |
3.816 ± 0.027 | 8.437 ± 0.042 |
2.045 ± 0.023 | 5.472 ± 0.029 |
3.815 ± 0.04 | 9.682 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.886 ± 0.027 | 2.983 ± 0.024 |
4.767 ± 0.031 | 3.486 ± 0.023 |
5.957 ± 0.047 | 5.537 ± 0.032 |
5.646 ± 0.059 | 7.127 ± 0.041 |
1.353 ± 0.018 | 2.268 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
