
Lake Sarah-associated circular virus-10
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.88
Get precalculated fractions of proteins
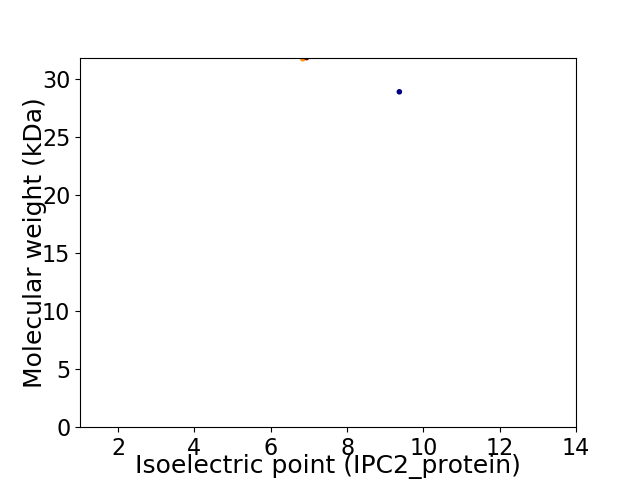
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
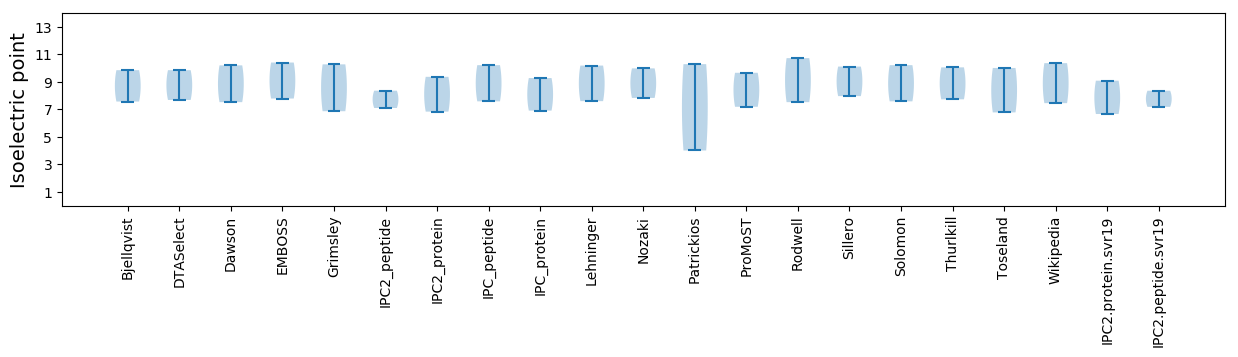
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQK3|A0A140AQK3_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular virus-10 OX=1685736 PE=3 SV=1
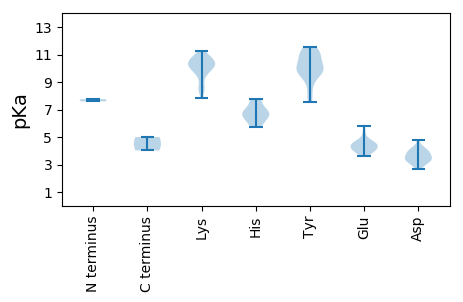
MM1 pKa = 7.75TDD3 pKa = 2.65TRR5 pKa = 11.84SRR7 pKa = 11.84DD8 pKa = 2.85WCFTLNNYY16 pKa = 7.52TEE18 pKa = 4.18EE19 pKa = 4.64EE20 pKa = 4.28YY21 pKa = 11.16DD22 pKa = 3.98VIHH25 pKa = 6.51SLSLSQYY32 pKa = 9.74KK33 pKa = 10.27YY34 pKa = 9.25IVVGKK39 pKa = 9.62EE40 pKa = 3.62VGEE43 pKa = 4.34SGTNHH48 pKa = 5.71LQGYY52 pKa = 9.67IYY54 pKa = 10.19FVNAKK59 pKa = 10.21SMSAVKK65 pKa = 10.61KK66 pKa = 9.48MISKK70 pKa = 10.01RR71 pKa = 11.84CHH73 pKa = 6.31LEE75 pKa = 3.77SAKK78 pKa = 10.57GSPLQAATYY87 pKa = 9.6CKK89 pKa = 10.16KK90 pKa = 10.83DD91 pKa = 3.37NNDD94 pKa = 3.24YY95 pKa = 11.12YY96 pKa = 11.17EE97 pKa = 4.43NGEE100 pKa = 4.14LPVIQGKK107 pKa = 7.81RR108 pKa = 11.84TDD110 pKa = 3.66LDD112 pKa = 4.02EE113 pKa = 5.28IRR115 pKa = 11.84DD116 pKa = 3.76ILKK119 pKa = 8.42QTNKK123 pKa = 9.47MSDD126 pKa = 3.09VVMVAKK132 pKa = 10.41SYY134 pKa = 11.07QSVKK138 pKa = 9.87MAEE141 pKa = 4.24QILKK145 pKa = 8.97YY146 pKa = 10.0HH147 pKa = 6.79EE148 pKa = 4.48KK149 pKa = 10.4PRR151 pKa = 11.84MEE153 pKa = 4.1KK154 pKa = 10.25PYY156 pKa = 10.51VEE158 pKa = 4.59WYY160 pKa = 9.99YY161 pKa = 11.56GPTGTGKK168 pKa = 10.18SKK170 pKa = 10.63KK171 pKa = 9.86AYY173 pKa = 8.7EE174 pKa = 4.13VLSDD178 pKa = 3.52EE179 pKa = 5.83CYY181 pKa = 9.81TCLSTGKK188 pKa = 9.52WFDD191 pKa = 3.95GYY193 pKa = 10.82DD194 pKa = 2.99AHH196 pKa = 7.74KK197 pKa = 10.91NVLIDD202 pKa = 4.76DD203 pKa = 3.82MRR205 pKa = 11.84KK206 pKa = 10.49DD207 pKa = 3.54FMKK210 pKa = 10.43FHH212 pKa = 6.83EE213 pKa = 4.91LLRR216 pKa = 11.84LLDD219 pKa = 4.22RR220 pKa = 11.84YY221 pKa = 11.07AMRR224 pKa = 11.84VEE226 pKa = 4.5CKK228 pKa = 10.11GGTRR232 pKa = 11.84QFVASHH238 pKa = 6.9IIITSCYY245 pKa = 8.27HH246 pKa = 6.1PKK248 pKa = 11.19DD249 pKa = 3.43MFEE252 pKa = 4.03TRR254 pKa = 11.84EE255 pKa = 4.67DD256 pKa = 3.29IQQLLRR262 pKa = 11.84RR263 pKa = 11.84IDD265 pKa = 3.7KK266 pKa = 10.63IEE268 pKa = 3.96NFEE271 pKa = 4.03
MM1 pKa = 7.75TDD3 pKa = 2.65TRR5 pKa = 11.84SRR7 pKa = 11.84DD8 pKa = 2.85WCFTLNNYY16 pKa = 7.52TEE18 pKa = 4.18EE19 pKa = 4.64EE20 pKa = 4.28YY21 pKa = 11.16DD22 pKa = 3.98VIHH25 pKa = 6.51SLSLSQYY32 pKa = 9.74KK33 pKa = 10.27YY34 pKa = 9.25IVVGKK39 pKa = 9.62EE40 pKa = 3.62VGEE43 pKa = 4.34SGTNHH48 pKa = 5.71LQGYY52 pKa = 9.67IYY54 pKa = 10.19FVNAKK59 pKa = 10.21SMSAVKK65 pKa = 10.61KK66 pKa = 9.48MISKK70 pKa = 10.01RR71 pKa = 11.84CHH73 pKa = 6.31LEE75 pKa = 3.77SAKK78 pKa = 10.57GSPLQAATYY87 pKa = 9.6CKK89 pKa = 10.16KK90 pKa = 10.83DD91 pKa = 3.37NNDD94 pKa = 3.24YY95 pKa = 11.12YY96 pKa = 11.17EE97 pKa = 4.43NGEE100 pKa = 4.14LPVIQGKK107 pKa = 7.81RR108 pKa = 11.84TDD110 pKa = 3.66LDD112 pKa = 4.02EE113 pKa = 5.28IRR115 pKa = 11.84DD116 pKa = 3.76ILKK119 pKa = 8.42QTNKK123 pKa = 9.47MSDD126 pKa = 3.09VVMVAKK132 pKa = 10.41SYY134 pKa = 11.07QSVKK138 pKa = 9.87MAEE141 pKa = 4.24QILKK145 pKa = 8.97YY146 pKa = 10.0HH147 pKa = 6.79EE148 pKa = 4.48KK149 pKa = 10.4PRR151 pKa = 11.84MEE153 pKa = 4.1KK154 pKa = 10.25PYY156 pKa = 10.51VEE158 pKa = 4.59WYY160 pKa = 9.99YY161 pKa = 11.56GPTGTGKK168 pKa = 10.18SKK170 pKa = 10.63KK171 pKa = 9.86AYY173 pKa = 8.7EE174 pKa = 4.13VLSDD178 pKa = 3.52EE179 pKa = 5.83CYY181 pKa = 9.81TCLSTGKK188 pKa = 9.52WFDD191 pKa = 3.95GYY193 pKa = 10.82DD194 pKa = 2.99AHH196 pKa = 7.74KK197 pKa = 10.91NVLIDD202 pKa = 4.76DD203 pKa = 3.82MRR205 pKa = 11.84KK206 pKa = 10.49DD207 pKa = 3.54FMKK210 pKa = 10.43FHH212 pKa = 6.83EE213 pKa = 4.91LLRR216 pKa = 11.84LLDD219 pKa = 4.22RR220 pKa = 11.84YY221 pKa = 11.07AMRR224 pKa = 11.84VEE226 pKa = 4.5CKK228 pKa = 10.11GGTRR232 pKa = 11.84QFVASHH238 pKa = 6.9IIITSCYY245 pKa = 8.27HH246 pKa = 6.1PKK248 pKa = 11.19DD249 pKa = 3.43MFEE252 pKa = 4.03TRR254 pKa = 11.84EE255 pKa = 4.67DD256 pKa = 3.29IQQLLRR262 pKa = 11.84RR263 pKa = 11.84IDD265 pKa = 3.7KK266 pKa = 10.63IEE268 pKa = 3.96NFEE271 pKa = 4.03
Molecular weight: 31.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQK3|A0A140AQK3_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular virus-10 OX=1685736 PE=3 SV=1
MM1 pKa = 7.59AKK3 pKa = 9.96FVKK6 pKa = 10.17KK7 pKa = 10.21PYY9 pKa = 9.93RR10 pKa = 11.84KK11 pKa = 8.78VRR13 pKa = 11.84KK14 pKa = 8.7YY15 pKa = 10.87AKK17 pKa = 10.24KK18 pKa = 9.7PVKK21 pKa = 10.64KK22 pKa = 9.97MMKK25 pKa = 8.35TSKK28 pKa = 10.73SFVKK32 pKa = 10.17KK33 pKa = 8.31VQKK36 pKa = 10.25IIHH39 pKa = 6.8KK40 pKa = 10.14DD41 pKa = 3.25VEE43 pKa = 4.58TKK45 pKa = 10.79VVVFNSNATAFNQQINSTGDD65 pKa = 3.63CLRR68 pKa = 11.84LLPDD72 pKa = 3.58IVNGTSEE79 pKa = 3.91NTKK82 pKa = 9.77IGNIIQLQSLNIRR95 pKa = 11.84GVLTFALSQTASQNVRR111 pKa = 11.84IGVRR115 pKa = 11.84MLILRR120 pKa = 11.84AKK122 pKa = 10.34RR123 pKa = 11.84FNDD126 pKa = 3.19WNQSATDD133 pKa = 3.92FATNYY138 pKa = 7.65TKK140 pKa = 10.83LLEE143 pKa = 4.65GSTSGFDD150 pKa = 3.44GSVAAFNTPVNHH162 pKa = 7.72DD163 pKa = 4.07YY164 pKa = 11.19FSVVADD170 pKa = 3.26KK171 pKa = 10.78RR172 pKa = 11.84FYY174 pKa = 10.07MSQSVILASGPTINTNEE191 pKa = 3.88TTKK194 pKa = 10.69FINFSVPYY202 pKa = 9.66SRR204 pKa = 11.84RR205 pKa = 11.84KK206 pKa = 8.51LTYY209 pKa = 9.87DD210 pKa = 3.0QDD212 pKa = 4.26FSGLAPTNYY221 pKa = 9.49PYY223 pKa = 11.28FMVLGYY229 pKa = 10.61SKK231 pKa = 11.24LDD233 pKa = 3.36GSVADD238 pKa = 4.31GTGTTYY244 pKa = 10.12LTFQYY249 pKa = 9.98TATAKK254 pKa = 10.91FEE256 pKa = 4.38DD257 pKa = 3.92AA258 pKa = 5.0
MM1 pKa = 7.59AKK3 pKa = 9.96FVKK6 pKa = 10.17KK7 pKa = 10.21PYY9 pKa = 9.93RR10 pKa = 11.84KK11 pKa = 8.78VRR13 pKa = 11.84KK14 pKa = 8.7YY15 pKa = 10.87AKK17 pKa = 10.24KK18 pKa = 9.7PVKK21 pKa = 10.64KK22 pKa = 9.97MMKK25 pKa = 8.35TSKK28 pKa = 10.73SFVKK32 pKa = 10.17KK33 pKa = 8.31VQKK36 pKa = 10.25IIHH39 pKa = 6.8KK40 pKa = 10.14DD41 pKa = 3.25VEE43 pKa = 4.58TKK45 pKa = 10.79VVVFNSNATAFNQQINSTGDD65 pKa = 3.63CLRR68 pKa = 11.84LLPDD72 pKa = 3.58IVNGTSEE79 pKa = 3.91NTKK82 pKa = 9.77IGNIIQLQSLNIRR95 pKa = 11.84GVLTFALSQTASQNVRR111 pKa = 11.84IGVRR115 pKa = 11.84MLILRR120 pKa = 11.84AKK122 pKa = 10.34RR123 pKa = 11.84FNDD126 pKa = 3.19WNQSATDD133 pKa = 3.92FATNYY138 pKa = 7.65TKK140 pKa = 10.83LLEE143 pKa = 4.65GSTSGFDD150 pKa = 3.44GSVAAFNTPVNHH162 pKa = 7.72DD163 pKa = 4.07YY164 pKa = 11.19FSVVADD170 pKa = 3.26KK171 pKa = 10.78RR172 pKa = 11.84FYY174 pKa = 10.07MSQSVILASGPTINTNEE191 pKa = 3.88TTKK194 pKa = 10.69FINFSVPYY202 pKa = 9.66SRR204 pKa = 11.84RR205 pKa = 11.84KK206 pKa = 8.51LTYY209 pKa = 9.87DD210 pKa = 3.0QDD212 pKa = 4.26FSGLAPTNYY221 pKa = 9.49PYY223 pKa = 11.28FMVLGYY229 pKa = 10.61SKK231 pKa = 11.24LDD233 pKa = 3.36GSVADD238 pKa = 4.31GTGTTYY244 pKa = 10.12LTFQYY249 pKa = 9.98TATAKK254 pKa = 10.91FEE256 pKa = 4.38DD257 pKa = 3.92AA258 pKa = 5.0
Molecular weight: 28.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
529 |
258 |
271 |
264.5 |
30.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.482 ± 0.967 | 1.512 ± 0.728 |
6.049 ± 0.654 | 4.915 ± 1.927 |
4.726 ± 1.206 | 5.293 ± 0.086 |
1.89 ± 0.722 | 5.293 ± 0.165 |
9.641 ± 0.219 | 6.805 ± 0.14 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.214 ± 0.575 | 5.293 ± 1.09 |
2.647 ± 0.294 | 3.97 ± 0.19 |
4.726 ± 0.299 | 7.372 ± 0.496 |
7.561 ± 1.377 | 6.994 ± 0.741 |
0.756 ± 0.239 | 5.86 ± 0.782 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
