
Pacific flying fox faeces associated gemycircularvirus-10
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus ptero1; Pteropus associated gemycircularvirus 1
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
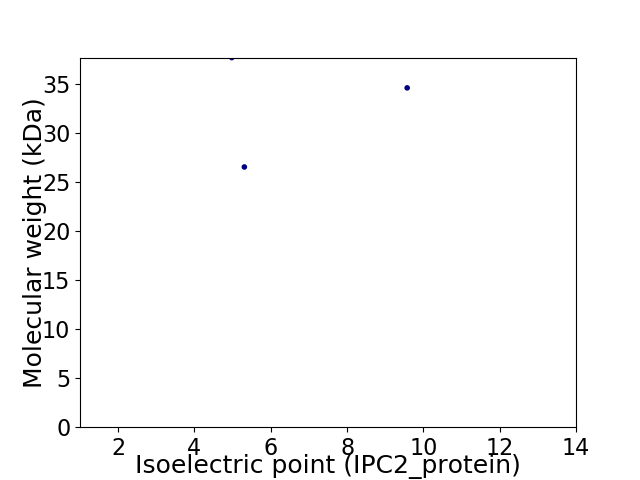
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
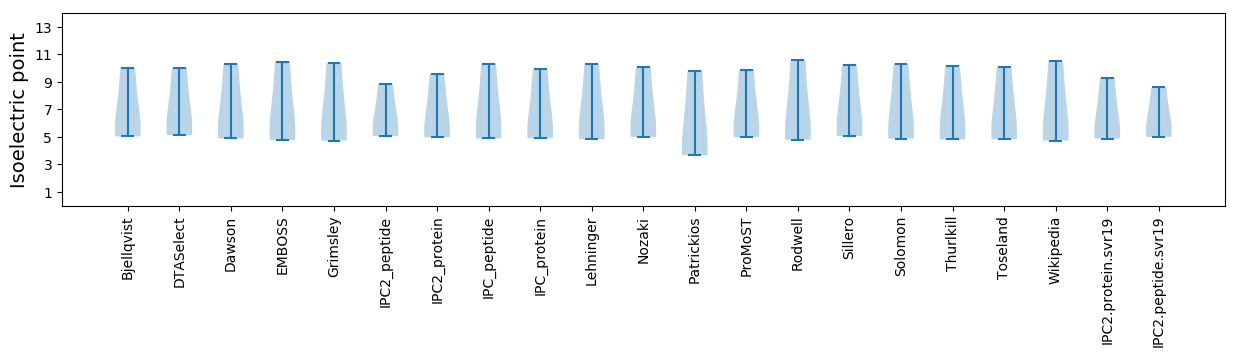
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTP6|A0A140CTP6_9VIRU RepA OS=Pacific flying fox faeces associated gemycircularvirus-10 OX=1795989 PE=3 SV=1
MM1 pKa = 7.67PLAVNAQHH9 pKa = 7.09FLLTYY14 pKa = 10.77AHH16 pKa = 6.8VEE18 pKa = 4.23GNDD21 pKa = 4.12DD22 pKa = 4.83KK23 pKa = 11.84PEE25 pKa = 4.66LDD27 pKa = 3.51PFRR30 pKa = 11.84IVDD33 pKa = 3.47VLGAKK38 pKa = 9.68GAEE41 pKa = 4.05CIVARR46 pKa = 11.84EE47 pKa = 4.39FYY49 pKa = 9.97PNTGGFHH56 pKa = 5.74FHH58 pKa = 6.18VFCSFEE64 pKa = 4.49RR65 pKa = 11.84NFRR68 pKa = 11.84SRR70 pKa = 11.84KK71 pKa = 9.65ADD73 pKa = 3.32VFDD76 pKa = 3.53VDD78 pKa = 5.1GYY80 pKa = 10.96HH81 pKa = 7.21PNIEE85 pKa = 4.12PSRR88 pKa = 11.84KK89 pKa = 8.63NAAGGFDD96 pKa = 4.07YY97 pKa = 9.3ATKK100 pKa = 10.78DD101 pKa = 3.18GDD103 pKa = 3.7IVAGGLEE110 pKa = 4.28RR111 pKa = 11.84PSRR114 pKa = 11.84TRR116 pKa = 11.84GRR118 pKa = 11.84AGTTGAQATWGEE130 pKa = 3.89ITAAEE135 pKa = 4.18SAEE138 pKa = 4.28EE139 pKa = 4.01FWRR142 pKa = 11.84LCEE145 pKa = 3.96EE146 pKa = 4.93LDD148 pKa = 3.94PKK150 pKa = 11.64SMVCNFPALSKK161 pKa = 9.48FAEE164 pKa = 4.1WRR166 pKa = 11.84FRR168 pKa = 11.84PIPVPYY174 pKa = 10.04ASPDD178 pKa = 3.48GVFDD182 pKa = 3.56TSGYY186 pKa = 9.74PIIEE190 pKa = 4.28EE191 pKa = 4.04WRR193 pKa = 11.84SSVFDD198 pKa = 4.0EE199 pKa = 4.2YY200 pKa = 11.35SGRR203 pKa = 11.84GEE205 pKa = 4.34SYY207 pKa = 10.05TYY209 pKa = 9.51LTSLVLFGPTRR220 pKa = 11.84LGKK223 pKa = 6.32TTWARR228 pKa = 11.84SLGSHH233 pKa = 6.88IYY235 pKa = 10.5FGGLFSAGEE244 pKa = 3.96AMRR247 pKa = 11.84CPEE250 pKa = 4.14AEE252 pKa = 4.08YY253 pKa = 10.59AIMDD257 pKa = 5.62DD258 pKa = 3.07IAGGIKK264 pKa = 10.03FFPRR268 pKa = 11.84FKK270 pKa = 10.86DD271 pKa = 3.16WLGCQAQFQLKK282 pKa = 9.74VLYY285 pKa = 10.02KK286 pKa = 10.74EE287 pKa = 3.79PALYY291 pKa = 10.33DD292 pKa = 3.25WGKK295 pKa = 9.31PCIWCYY301 pKa = 10.84NVDD304 pKa = 3.46PRR306 pKa = 11.84AGMSVEE312 pKa = 4.32DD313 pKa = 4.27IEE315 pKa = 4.34WLEE318 pKa = 4.17GNCVFVEE325 pKa = 3.98ITSPIFHH332 pKa = 7.38ANTEE336 pKa = 4.14
MM1 pKa = 7.67PLAVNAQHH9 pKa = 7.09FLLTYY14 pKa = 10.77AHH16 pKa = 6.8VEE18 pKa = 4.23GNDD21 pKa = 4.12DD22 pKa = 4.83KK23 pKa = 11.84PEE25 pKa = 4.66LDD27 pKa = 3.51PFRR30 pKa = 11.84IVDD33 pKa = 3.47VLGAKK38 pKa = 9.68GAEE41 pKa = 4.05CIVARR46 pKa = 11.84EE47 pKa = 4.39FYY49 pKa = 9.97PNTGGFHH56 pKa = 5.74FHH58 pKa = 6.18VFCSFEE64 pKa = 4.49RR65 pKa = 11.84NFRR68 pKa = 11.84SRR70 pKa = 11.84KK71 pKa = 9.65ADD73 pKa = 3.32VFDD76 pKa = 3.53VDD78 pKa = 5.1GYY80 pKa = 10.96HH81 pKa = 7.21PNIEE85 pKa = 4.12PSRR88 pKa = 11.84KK89 pKa = 8.63NAAGGFDD96 pKa = 4.07YY97 pKa = 9.3ATKK100 pKa = 10.78DD101 pKa = 3.18GDD103 pKa = 3.7IVAGGLEE110 pKa = 4.28RR111 pKa = 11.84PSRR114 pKa = 11.84TRR116 pKa = 11.84GRR118 pKa = 11.84AGTTGAQATWGEE130 pKa = 3.89ITAAEE135 pKa = 4.18SAEE138 pKa = 4.28EE139 pKa = 4.01FWRR142 pKa = 11.84LCEE145 pKa = 3.96EE146 pKa = 4.93LDD148 pKa = 3.94PKK150 pKa = 11.64SMVCNFPALSKK161 pKa = 9.48FAEE164 pKa = 4.1WRR166 pKa = 11.84FRR168 pKa = 11.84PIPVPYY174 pKa = 10.04ASPDD178 pKa = 3.48GVFDD182 pKa = 3.56TSGYY186 pKa = 9.74PIIEE190 pKa = 4.28EE191 pKa = 4.04WRR193 pKa = 11.84SSVFDD198 pKa = 4.0EE199 pKa = 4.2YY200 pKa = 11.35SGRR203 pKa = 11.84GEE205 pKa = 4.34SYY207 pKa = 10.05TYY209 pKa = 9.51LTSLVLFGPTRR220 pKa = 11.84LGKK223 pKa = 6.32TTWARR228 pKa = 11.84SLGSHH233 pKa = 6.88IYY235 pKa = 10.5FGGLFSAGEE244 pKa = 3.96AMRR247 pKa = 11.84CPEE250 pKa = 4.14AEE252 pKa = 4.08YY253 pKa = 10.59AIMDD257 pKa = 5.62DD258 pKa = 3.07IAGGIKK264 pKa = 10.03FFPRR268 pKa = 11.84FKK270 pKa = 10.86DD271 pKa = 3.16WLGCQAQFQLKK282 pKa = 9.74VLYY285 pKa = 10.02KK286 pKa = 10.74EE287 pKa = 3.79PALYY291 pKa = 10.33DD292 pKa = 3.25WGKK295 pKa = 9.31PCIWCYY301 pKa = 10.84NVDD304 pKa = 3.46PRR306 pKa = 11.84AGMSVEE312 pKa = 4.32DD313 pKa = 4.27IEE315 pKa = 4.34WLEE318 pKa = 4.17GNCVFVEE325 pKa = 3.98ITSPIFHH332 pKa = 7.38ANTEE336 pKa = 4.14
Molecular weight: 37.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTP5|A0A140CTP5_9VIRU Replication-associated protein OS=Pacific flying fox faeces associated gemycircularvirus-10 OX=1795989 PE=3 SV=1
MM1 pKa = 7.87AYY3 pKa = 10.08ARR5 pKa = 11.84SSTRR9 pKa = 11.84RR10 pKa = 11.84SYY12 pKa = 10.85RR13 pKa = 11.84RR14 pKa = 11.84SSRR17 pKa = 11.84YY18 pKa = 8.8ARR20 pKa = 11.84RR21 pKa = 11.84KK22 pKa = 8.87IPHH25 pKa = 4.55TRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84YY30 pKa = 8.87VKK32 pKa = 8.37RR33 pKa = 11.84TSRR36 pKa = 11.84TPYY39 pKa = 8.78RR40 pKa = 11.84RR41 pKa = 11.84KK42 pKa = 8.27MTQKK46 pKa = 10.65KK47 pKa = 9.43ILNLTSIKK55 pKa = 10.5KK56 pKa = 9.71RR57 pKa = 11.84DD58 pKa = 3.61TMLSLTNVIADD69 pKa = 4.14RR70 pKa = 11.84SRR72 pKa = 11.84PTSYY76 pKa = 10.5TSNDD80 pKa = 3.02AVLSGGAQPYY90 pKa = 9.23IIPWVATARR99 pKa = 11.84TTTVNGTKK107 pKa = 9.95QGTIATSATRR117 pKa = 11.84SSTTCYY123 pKa = 10.16IRR125 pKa = 11.84GLRR128 pKa = 11.84EE129 pKa = 4.3DD130 pKa = 3.39IEE132 pKa = 4.78INVFTGCPWQWRR144 pKa = 11.84RR145 pKa = 11.84IVFFSKK151 pKa = 10.79GLTNAVGTPGSNFYY165 pKa = 10.43YY166 pKa = 10.78FNVTAEE172 pKa = 4.26DD173 pKa = 3.57YY174 pKa = 10.72RR175 pKa = 11.84RR176 pKa = 11.84TVNEE180 pKa = 3.7IYY182 pKa = 10.51GAYY185 pKa = 10.44ANGLNAFLFKK195 pKa = 10.25GTEE198 pKa = 4.03GSDD201 pKa = 3.11WNDD204 pKa = 3.1LMIAPVDD211 pKa = 3.67TTRR214 pKa = 11.84VSVLYY219 pKa = 10.78DD220 pKa = 3.16KK221 pKa = 10.97TRR223 pKa = 11.84TIASGNEE230 pKa = 3.37EE231 pKa = 4.02GVIRR235 pKa = 11.84KK236 pKa = 9.39YY237 pKa = 11.38KK238 pKa = 8.64MWHH241 pKa = 5.6GVNKK245 pKa = 10.27NLTYY249 pKa = 11.09DD250 pKa = 3.81DD251 pKa = 5.41DD252 pKa = 4.42EE253 pKa = 5.82FGGDD257 pKa = 3.33EE258 pKa = 4.33TTSNLSTLSKK268 pKa = 10.5VGCGDD273 pKa = 3.41MMVVDD278 pKa = 3.81IFKK281 pKa = 10.46PRR283 pKa = 11.84GGSAGEE289 pKa = 3.88DD290 pKa = 2.51RR291 pKa = 11.84MAFKK295 pKa = 10.8PNATLYY301 pKa = 8.81WHH303 pKa = 6.99EE304 pKa = 4.27KK305 pKa = 9.3
MM1 pKa = 7.87AYY3 pKa = 10.08ARR5 pKa = 11.84SSTRR9 pKa = 11.84RR10 pKa = 11.84SYY12 pKa = 10.85RR13 pKa = 11.84RR14 pKa = 11.84SSRR17 pKa = 11.84YY18 pKa = 8.8ARR20 pKa = 11.84RR21 pKa = 11.84KK22 pKa = 8.87IPHH25 pKa = 4.55TRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84YY30 pKa = 8.87VKK32 pKa = 8.37RR33 pKa = 11.84TSRR36 pKa = 11.84TPYY39 pKa = 8.78RR40 pKa = 11.84RR41 pKa = 11.84KK42 pKa = 8.27MTQKK46 pKa = 10.65KK47 pKa = 9.43ILNLTSIKK55 pKa = 10.5KK56 pKa = 9.71RR57 pKa = 11.84DD58 pKa = 3.61TMLSLTNVIADD69 pKa = 4.14RR70 pKa = 11.84SRR72 pKa = 11.84PTSYY76 pKa = 10.5TSNDD80 pKa = 3.02AVLSGGAQPYY90 pKa = 9.23IIPWVATARR99 pKa = 11.84TTTVNGTKK107 pKa = 9.95QGTIATSATRR117 pKa = 11.84SSTTCYY123 pKa = 10.16IRR125 pKa = 11.84GLRR128 pKa = 11.84EE129 pKa = 4.3DD130 pKa = 3.39IEE132 pKa = 4.78INVFTGCPWQWRR144 pKa = 11.84RR145 pKa = 11.84IVFFSKK151 pKa = 10.79GLTNAVGTPGSNFYY165 pKa = 10.43YY166 pKa = 10.78FNVTAEE172 pKa = 4.26DD173 pKa = 3.57YY174 pKa = 10.72RR175 pKa = 11.84RR176 pKa = 11.84TVNEE180 pKa = 3.7IYY182 pKa = 10.51GAYY185 pKa = 10.44ANGLNAFLFKK195 pKa = 10.25GTEE198 pKa = 4.03GSDD201 pKa = 3.11WNDD204 pKa = 3.1LMIAPVDD211 pKa = 3.67TTRR214 pKa = 11.84VSVLYY219 pKa = 10.78DD220 pKa = 3.16KK221 pKa = 10.97TRR223 pKa = 11.84TIASGNEE230 pKa = 3.37EE231 pKa = 4.02GVIRR235 pKa = 11.84KK236 pKa = 9.39YY237 pKa = 11.38KK238 pKa = 8.64MWHH241 pKa = 5.6GVNKK245 pKa = 10.27NLTYY249 pKa = 11.09DD250 pKa = 3.81DD251 pKa = 5.41DD252 pKa = 4.42EE253 pKa = 5.82FGGDD257 pKa = 3.33EE258 pKa = 4.33TTSNLSTLSKK268 pKa = 10.5VGCGDD273 pKa = 3.41MMVVDD278 pKa = 3.81IFKK281 pKa = 10.46PRR283 pKa = 11.84GGSAGEE289 pKa = 3.88DD290 pKa = 2.51RR291 pKa = 11.84MAFKK295 pKa = 10.8PNATLYY301 pKa = 8.81WHH303 pKa = 6.99EE304 pKa = 4.27KK305 pKa = 9.3
Molecular weight: 34.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
879 |
238 |
336 |
293.0 |
32.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.191 ± 0.665 | 1.934 ± 0.403 |
5.688 ± 0.18 | 6.371 ± 1.125 |
6.143 ± 1.166 | 8.532 ± 0.41 |
1.82 ± 0.351 | 4.551 ± 0.404 |
4.323 ± 0.668 | 5.575 ± 0.266 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.706 ± 0.395 | 3.868 ± 0.691 |
5.575 ± 0.969 | 1.365 ± 0.052 |
7.85 ± 0.995 | 6.598 ± 0.532 |
7.395 ± 1.67 | 5.802 ± 0.062 |
2.162 ± 0.221 | 4.551 ± 0.422 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
