
Shewanella algae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Shewanellaceae; Shewanella
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
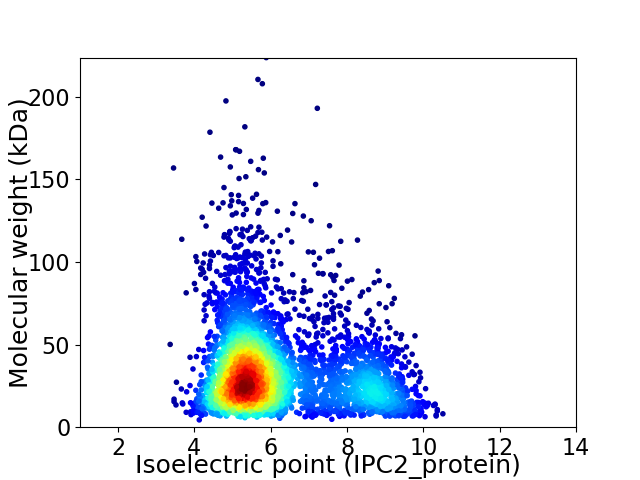
Virtual 2D-PAGE plot for 3935 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S2T8I8|A0A1S2T8I8_9GAMM Flagella-associated GTP-binding protein OS=Shewanella algae OX=38313 GN=BFS86_03580 PE=3 SV=1
MM1 pKa = 7.92KK2 pKa = 10.55LFQCNWLIRR11 pKa = 11.84ALGVVLIGGASLYY24 pKa = 10.29GASSLTPVNASFFPQQFDD42 pKa = 3.68QACMAEE48 pKa = 4.17QAGFALNCTANDD60 pKa = 3.45VRR62 pKa = 11.84VSKK65 pKa = 10.94VDD67 pKa = 3.26NVRR70 pKa = 11.84NLDD73 pKa = 3.19GTTPVEE79 pKa = 4.42CVLGDD84 pKa = 3.99DD85 pKa = 3.94VTFLADD91 pKa = 3.41VTITTTANQRR101 pKa = 11.84YY102 pKa = 9.03DD103 pKa = 3.48YY104 pKa = 11.06SVYY107 pKa = 10.48LPEE110 pKa = 5.14GSWSAQDD117 pKa = 3.46SDD119 pKa = 4.57PNNEE123 pKa = 3.81CSILLGRR130 pKa = 11.84TNGPGVDD137 pKa = 3.77LEE139 pKa = 4.64EE140 pKa = 4.98NLDD143 pKa = 3.7ACADD147 pKa = 3.31ISKK150 pKa = 10.47AAGYY154 pKa = 9.05DD155 pKa = 3.29ATHH158 pKa = 6.65VYY160 pKa = 10.35SGEE163 pKa = 4.13QITLFCRR170 pKa = 11.84DD171 pKa = 4.26DD172 pKa = 4.47DD173 pKa = 4.43NSGKK177 pKa = 10.66AEE179 pKa = 4.04FNYY182 pKa = 9.59CAAWHH187 pKa = 6.28NKK189 pKa = 7.81TGADD193 pKa = 3.32CSEE196 pKa = 4.5NNPAAPGTPSKK207 pKa = 10.53CRR209 pKa = 11.84CDD211 pKa = 4.08AFDD214 pKa = 3.88IDD216 pKa = 5.61VFIKK220 pKa = 10.44PNPPVINKK228 pKa = 9.68KK229 pKa = 10.55LITSNTQNEE238 pKa = 4.23PGGTYY243 pKa = 9.02TFEE246 pKa = 5.24LSFNNPNAQTSLFITSLSDD265 pKa = 3.43EE266 pKa = 4.27VDD268 pKa = 2.87IGADD272 pKa = 3.45GTYY275 pKa = 8.89DD276 pKa = 3.1TSLNLWSAGGPVGASDD292 pKa = 3.93GVYY295 pKa = 10.35LKK297 pKa = 10.83SSNCTQPANGGEE309 pKa = 4.08ILPSGSYY316 pKa = 9.21SCQFTVHH323 pKa = 6.39IVDD326 pKa = 4.6RR327 pKa = 11.84DD328 pKa = 3.93LPDD331 pKa = 4.07LAPNIEE337 pKa = 4.87LYY339 pKa = 10.97DD340 pKa = 4.12DD341 pKa = 5.3LIKK344 pKa = 10.86LALEE348 pKa = 4.76DD349 pKa = 4.23KK350 pKa = 11.17NGDD353 pKa = 3.79DD354 pKa = 4.87VIDD357 pKa = 4.32SNSCAAVNGIDD368 pKa = 5.23GEE370 pKa = 4.43HH371 pKa = 7.06CSNVVQVQVEE381 pKa = 4.22NLNPSITVLKK391 pKa = 8.8TANPDD396 pKa = 3.34QVPEE400 pKa = 4.05SGGNVTYY407 pKa = 8.62TVRR410 pKa = 11.84VNNTAEE416 pKa = 4.86AYY418 pKa = 9.07DD419 pKa = 3.83SPLEE423 pKa = 4.0LFYY426 pKa = 11.34LVDD429 pKa = 5.34DD430 pKa = 5.15KK431 pKa = 11.75FGDD434 pKa = 3.93LNGKK438 pKa = 8.81GSCSTYY444 pKa = 10.98LDD446 pKa = 3.27IAYY449 pKa = 9.93GSYY452 pKa = 10.25YY453 pKa = 9.79EE454 pKa = 4.51CSFTEE459 pKa = 5.36FISGTGAGSHH469 pKa = 6.02TNTVTAKK476 pKa = 10.54AYY478 pKa = 9.82DD479 pKa = 3.77DD480 pKa = 3.87EE481 pKa = 5.05MIEE484 pKa = 5.13AMAADD489 pKa = 3.94SATVVINDD497 pKa = 3.75IPSMITLDD505 pKa = 3.57KK506 pKa = 11.19SADD509 pKa = 3.61PTSVLEE515 pKa = 4.32TGDD518 pKa = 3.85DD519 pKa = 3.57LSIFRR524 pKa = 11.84DD525 pKa = 3.63VEE527 pKa = 4.22FTFLFGVKK535 pKa = 10.1DD536 pKa = 3.7QINGQNTVDD545 pKa = 3.06TVTFNTLDD553 pKa = 3.63DD554 pKa = 4.28SVFGDD559 pKa = 3.99LTAKK563 pKa = 10.81CEE565 pKa = 3.83VDD567 pKa = 3.57TVDD570 pKa = 3.86GLPYY574 pKa = 10.64GPAPLAGLMLDD585 pKa = 3.89PGHH588 pKa = 6.37NASCTITLQVQGNRR602 pKa = 11.84TDD604 pKa = 2.47IHH606 pKa = 6.42TNLATIYY613 pKa = 9.26GTDD616 pKa = 3.5EE617 pKa = 4.71DD618 pKa = 4.53GQAVDD623 pKa = 4.97AMDD626 pKa = 4.94DD627 pKa = 3.5ATVTFTPGSPAVDD640 pKa = 3.25MDD642 pKa = 4.49FAASMLVVLGMHH654 pKa = 6.49NADD657 pKa = 3.49VNNANLTMLTAAGVDD672 pKa = 3.73VFPGADD678 pKa = 2.99MPGFKK683 pKa = 10.36LINSGGTFEE692 pKa = 4.14GVNYY696 pKa = 9.72GSCALNHH703 pKa = 6.23PFGYY707 pKa = 8.89TGSGTEE713 pKa = 4.9DD714 pKa = 3.82YY715 pKa = 10.78FCAFTIEE722 pKa = 4.51FKK724 pKa = 10.8PGLEE728 pKa = 4.06NTDD731 pKa = 4.24PIVFLGDD738 pKa = 3.59VIVKK742 pKa = 9.45LVNSKK747 pKa = 10.77GDD749 pKa = 3.31EE750 pKa = 4.1STADD754 pKa = 3.19VTIQVGTNEE763 pKa = 3.96NN764 pKa = 3.4
MM1 pKa = 7.92KK2 pKa = 10.55LFQCNWLIRR11 pKa = 11.84ALGVVLIGGASLYY24 pKa = 10.29GASSLTPVNASFFPQQFDD42 pKa = 3.68QACMAEE48 pKa = 4.17QAGFALNCTANDD60 pKa = 3.45VRR62 pKa = 11.84VSKK65 pKa = 10.94VDD67 pKa = 3.26NVRR70 pKa = 11.84NLDD73 pKa = 3.19GTTPVEE79 pKa = 4.42CVLGDD84 pKa = 3.99DD85 pKa = 3.94VTFLADD91 pKa = 3.41VTITTTANQRR101 pKa = 11.84YY102 pKa = 9.03DD103 pKa = 3.48YY104 pKa = 11.06SVYY107 pKa = 10.48LPEE110 pKa = 5.14GSWSAQDD117 pKa = 3.46SDD119 pKa = 4.57PNNEE123 pKa = 3.81CSILLGRR130 pKa = 11.84TNGPGVDD137 pKa = 3.77LEE139 pKa = 4.64EE140 pKa = 4.98NLDD143 pKa = 3.7ACADD147 pKa = 3.31ISKK150 pKa = 10.47AAGYY154 pKa = 9.05DD155 pKa = 3.29ATHH158 pKa = 6.65VYY160 pKa = 10.35SGEE163 pKa = 4.13QITLFCRR170 pKa = 11.84DD171 pKa = 4.26DD172 pKa = 4.47DD173 pKa = 4.43NSGKK177 pKa = 10.66AEE179 pKa = 4.04FNYY182 pKa = 9.59CAAWHH187 pKa = 6.28NKK189 pKa = 7.81TGADD193 pKa = 3.32CSEE196 pKa = 4.5NNPAAPGTPSKK207 pKa = 10.53CRR209 pKa = 11.84CDD211 pKa = 4.08AFDD214 pKa = 3.88IDD216 pKa = 5.61VFIKK220 pKa = 10.44PNPPVINKK228 pKa = 9.68KK229 pKa = 10.55LITSNTQNEE238 pKa = 4.23PGGTYY243 pKa = 9.02TFEE246 pKa = 5.24LSFNNPNAQTSLFITSLSDD265 pKa = 3.43EE266 pKa = 4.27VDD268 pKa = 2.87IGADD272 pKa = 3.45GTYY275 pKa = 8.89DD276 pKa = 3.1TSLNLWSAGGPVGASDD292 pKa = 3.93GVYY295 pKa = 10.35LKK297 pKa = 10.83SSNCTQPANGGEE309 pKa = 4.08ILPSGSYY316 pKa = 9.21SCQFTVHH323 pKa = 6.39IVDD326 pKa = 4.6RR327 pKa = 11.84DD328 pKa = 3.93LPDD331 pKa = 4.07LAPNIEE337 pKa = 4.87LYY339 pKa = 10.97DD340 pKa = 4.12DD341 pKa = 5.3LIKK344 pKa = 10.86LALEE348 pKa = 4.76DD349 pKa = 4.23KK350 pKa = 11.17NGDD353 pKa = 3.79DD354 pKa = 4.87VIDD357 pKa = 4.32SNSCAAVNGIDD368 pKa = 5.23GEE370 pKa = 4.43HH371 pKa = 7.06CSNVVQVQVEE381 pKa = 4.22NLNPSITVLKK391 pKa = 8.8TANPDD396 pKa = 3.34QVPEE400 pKa = 4.05SGGNVTYY407 pKa = 8.62TVRR410 pKa = 11.84VNNTAEE416 pKa = 4.86AYY418 pKa = 9.07DD419 pKa = 3.83SPLEE423 pKa = 4.0LFYY426 pKa = 11.34LVDD429 pKa = 5.34DD430 pKa = 5.15KK431 pKa = 11.75FGDD434 pKa = 3.93LNGKK438 pKa = 8.81GSCSTYY444 pKa = 10.98LDD446 pKa = 3.27IAYY449 pKa = 9.93GSYY452 pKa = 10.25YY453 pKa = 9.79EE454 pKa = 4.51CSFTEE459 pKa = 5.36FISGTGAGSHH469 pKa = 6.02TNTVTAKK476 pKa = 10.54AYY478 pKa = 9.82DD479 pKa = 3.77DD480 pKa = 3.87EE481 pKa = 5.05MIEE484 pKa = 5.13AMAADD489 pKa = 3.94SATVVINDD497 pKa = 3.75IPSMITLDD505 pKa = 3.57KK506 pKa = 11.19SADD509 pKa = 3.61PTSVLEE515 pKa = 4.32TGDD518 pKa = 3.85DD519 pKa = 3.57LSIFRR524 pKa = 11.84DD525 pKa = 3.63VEE527 pKa = 4.22FTFLFGVKK535 pKa = 10.1DD536 pKa = 3.7QINGQNTVDD545 pKa = 3.06TVTFNTLDD553 pKa = 3.63DD554 pKa = 4.28SVFGDD559 pKa = 3.99LTAKK563 pKa = 10.81CEE565 pKa = 3.83VDD567 pKa = 3.57TVDD570 pKa = 3.86GLPYY574 pKa = 10.64GPAPLAGLMLDD585 pKa = 3.89PGHH588 pKa = 6.37NASCTITLQVQGNRR602 pKa = 11.84TDD604 pKa = 2.47IHH606 pKa = 6.42TNLATIYY613 pKa = 9.26GTDD616 pKa = 3.5EE617 pKa = 4.71DD618 pKa = 4.53GQAVDD623 pKa = 4.97AMDD626 pKa = 4.94DD627 pKa = 3.5ATVTFTPGSPAVDD640 pKa = 3.25MDD642 pKa = 4.49FAASMLVVLGMHH654 pKa = 6.49NADD657 pKa = 3.49VNNANLTMLTAAGVDD672 pKa = 3.73VFPGADD678 pKa = 2.99MPGFKK683 pKa = 10.36LINSGGTFEE692 pKa = 4.14GVNYY696 pKa = 9.72GSCALNHH703 pKa = 6.23PFGYY707 pKa = 8.89TGSGTEE713 pKa = 4.9DD714 pKa = 3.82YY715 pKa = 10.78FCAFTIEE722 pKa = 4.51FKK724 pKa = 10.8PGLEE728 pKa = 4.06NTDD731 pKa = 4.24PIVFLGDD738 pKa = 3.59VIVKK742 pKa = 9.45LVNSKK747 pKa = 10.77GDD749 pKa = 3.31EE750 pKa = 4.1STADD754 pKa = 3.19VTIQVGTNEE763 pKa = 3.96NN764 pKa = 3.4
Molecular weight: 81.26 kDa
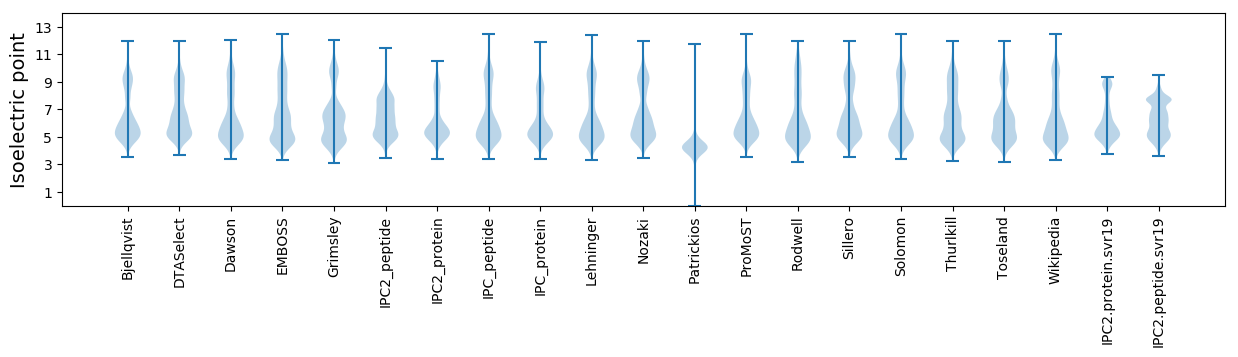
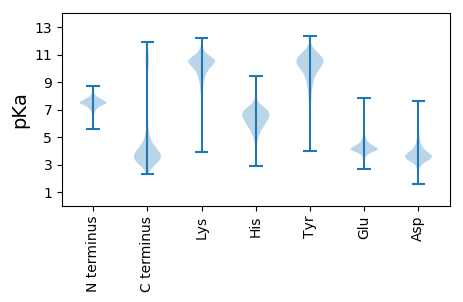
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S2TVI2|A0A1S2TVI2_9GAMM Fe-S assembly protein IscX OS=Shewanella algae OX=38313 GN=BFS86_12180 PE=4 SV=1
MM1 pKa = 7.69KK2 pKa = 10.31SLSLLQFPRR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 5.61PQALTIISRR22 pKa = 11.84ALASIIGGYY31 pKa = 10.17LIASLACALLAVSLPMSRR49 pKa = 11.84IDD51 pKa = 3.61STVCAMMLSFLIYY64 pKa = 9.01ALVVIRR70 pKa = 11.84VFCIKK75 pKa = 10.25RR76 pKa = 11.84SLRR79 pKa = 11.84AWQEE83 pKa = 3.89LLLLAAALFGVLKK96 pKa = 10.86LLGAA100 pKa = 4.7
MM1 pKa = 7.69KK2 pKa = 10.31SLSLLQFPRR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 5.61PQALTIISRR22 pKa = 11.84ALASIIGGYY31 pKa = 10.17LIASLACALLAVSLPMSRR49 pKa = 11.84IDD51 pKa = 3.61STVCAMMLSFLIYY64 pKa = 9.01ALVVIRR70 pKa = 11.84VFCIKK75 pKa = 10.25RR76 pKa = 11.84SLRR79 pKa = 11.84AWQEE83 pKa = 3.89LLLLAAALFGVLKK96 pKa = 10.86LLGAA100 pKa = 4.7
Molecular weight: 10.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1298577 |
37 |
1980 |
330.0 |
36.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.846 ± 0.045 | 1.113 ± 0.016 |
5.358 ± 0.029 | 6.184 ± 0.035 |
3.831 ± 0.026 | 7.326 ± 0.034 |
2.147 ± 0.019 | 5.356 ± 0.03 |
4.725 ± 0.032 | 11.662 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.558 ± 0.02 | 3.58 ± 0.027 |
4.255 ± 0.025 | 5.06 ± 0.037 |
5.213 ± 0.033 | 6.366 ± 0.03 |
4.711 ± 0.027 | 6.456 ± 0.034 |
1.337 ± 0.018 | 2.917 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
