
Beihai astro-like virus
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
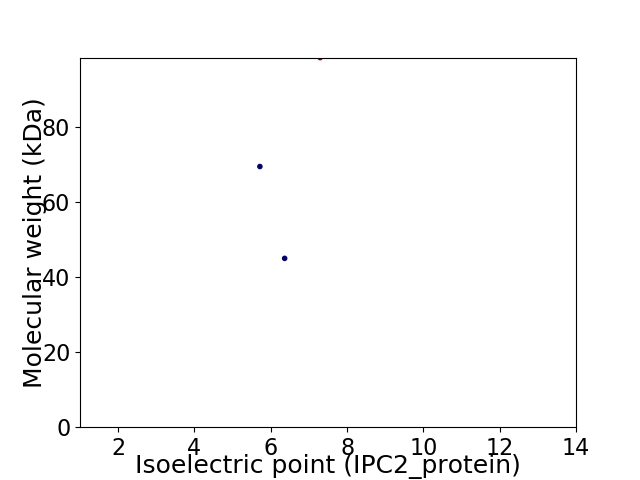
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KNT1|A0A1L3KNT1_9VIRU Putative capsid OS=Beihai astro-like virus OX=1922353 PE=4 SV=1
MM1 pKa = 7.74EE2 pKa = 4.98KK3 pKa = 9.65TPKK6 pKa = 9.07EE7 pKa = 3.79QRR9 pKa = 11.84TPRR12 pKa = 11.84VKK14 pKa = 10.37KK15 pKa = 9.36QQPQKK20 pKa = 10.86KK21 pKa = 8.83PAGQQQPRR29 pKa = 11.84KK30 pKa = 8.66PAQQKK35 pKa = 9.51RR36 pKa = 11.84RR37 pKa = 11.84SPMPQKK43 pKa = 9.18THH45 pKa = 6.97DD46 pKa = 3.67GEE48 pKa = 4.8IYY50 pKa = 10.41SSSSLVSIIHH60 pKa = 6.6GNQDD64 pKa = 4.01DD65 pKa = 4.42FTTPLAISLNPLVVAEE81 pKa = 4.17EE82 pKa = 4.18AKK84 pKa = 11.04DD85 pKa = 3.71PILNMKK91 pKa = 9.47CMSYY95 pKa = 11.13NCFKK99 pKa = 10.98VLSAKK104 pKa = 10.73LEE106 pKa = 4.18LKK108 pKa = 10.19AVCGPNAAVGTTATVGSWQNVDD130 pKa = 5.05DD131 pKa = 4.19PTGPQTPNTALKK143 pKa = 8.98MDD145 pKa = 4.11RR146 pKa = 11.84AVTVGIGMRR155 pKa = 11.84ACAPIKK161 pKa = 10.62HH162 pKa = 5.99SDD164 pKa = 3.55EE165 pKa = 4.34VKK167 pKa = 10.52VVYY170 pKa = 10.06TEE172 pKa = 3.79ATAAPRR178 pKa = 11.84TWNPGHH184 pKa = 7.12AIVSLIGQTGTVYY197 pKa = 10.6NQEE200 pKa = 3.84QTPVWKK206 pKa = 10.37RR207 pKa = 11.84PLWHH211 pKa = 7.05AFLHH215 pKa = 4.81YY216 pKa = 8.86TYY218 pKa = 10.58HH219 pKa = 5.56FTSPKK224 pKa = 10.12DD225 pKa = 3.39NRR227 pKa = 11.84NRR229 pKa = 11.84GSLAAGDD236 pKa = 5.09LDD238 pKa = 3.84DD239 pKa = 5.1QKK241 pKa = 11.71VSLHH245 pKa = 6.72EE246 pKa = 4.62IPDD249 pKa = 3.68EE250 pKa = 4.14PLVLEE255 pKa = 4.81MPNSPAARR263 pKa = 11.84KK264 pKa = 8.98LICRR268 pKa = 11.84GRR270 pKa = 11.84KK271 pKa = 7.69AQGRR275 pKa = 11.84GRR277 pKa = 11.84AGLWDD282 pKa = 4.16FASGILQGLSGVLPPPWGSLLNAGVALVRR311 pKa = 11.84PKK313 pKa = 10.08TATTRR318 pKa = 11.84EE319 pKa = 3.82GDD321 pKa = 3.87TIQLEE326 pKa = 4.39LFQTLSDD333 pKa = 3.88VAQDD337 pKa = 3.62QPVMGVEE344 pKa = 3.93ARR346 pKa = 11.84TTQMDD351 pKa = 5.76LINGHH356 pKa = 5.25YY357 pKa = 9.1TQMTDD362 pKa = 2.9PGANSVLEE370 pKa = 4.53DD371 pKa = 3.53SDD373 pKa = 4.05GGGGVLPPTGGGIVWHH389 pKa = 7.2DD390 pKa = 4.45ANPDD394 pKa = 3.32DD395 pKa = 4.6WSGDD399 pKa = 3.25EE400 pKa = 3.97QIIVNSYY407 pKa = 7.46VNKK410 pKa = 10.53YY411 pKa = 9.55ADD413 pKa = 3.21NYY415 pKa = 9.48FNEE418 pKa = 4.45VYY420 pKa = 9.87PVYY423 pKa = 10.68SSSDD427 pKa = 3.25VNYY430 pKa = 10.6NITVTKK436 pKa = 10.68GPLAGNNYY444 pKa = 8.69QFYY447 pKa = 10.46HH448 pKa = 7.71AMEE451 pKa = 4.18FQKK454 pKa = 10.16WKK456 pKa = 9.4HH457 pKa = 5.26QNTEE461 pKa = 3.81VTAVWVRR468 pKa = 11.84FQKK471 pKa = 9.98MSNMDD476 pKa = 3.95LQAKK480 pKa = 8.63GYY482 pKa = 10.4QMYY485 pKa = 10.5LPQFIEE491 pKa = 4.13KK492 pKa = 8.38FTNASSTSGEE502 pKa = 4.17EE503 pKa = 3.99KK504 pKa = 10.44FVMLAKK510 pKa = 10.73DD511 pKa = 3.5GGRR514 pKa = 11.84VQLQSKK520 pKa = 10.12SSPTVFTRR528 pKa = 11.84FLNMNSSTPWSALFLGEE545 pKa = 4.06SRR547 pKa = 11.84SIKK550 pKa = 9.84IYY552 pKa = 10.56RR553 pKa = 11.84DD554 pKa = 3.59GTSLAAGNWNPCQDD568 pKa = 3.78VLPGNNLSNLQATTAAAGDD587 pKa = 3.98EE588 pKa = 4.43EE589 pKa = 4.37EE590 pKa = 5.56AEE592 pKa = 4.05ADD594 pKa = 5.13KK595 pKa = 11.4EE596 pKa = 4.13DD597 pKa = 3.74LLDD600 pKa = 4.95RR601 pKa = 11.84IRR603 pKa = 11.84YY604 pKa = 8.77LEE606 pKa = 4.45RR607 pKa = 11.84EE608 pKa = 4.37LEE610 pKa = 4.2SAEE613 pKa = 4.14TEE615 pKa = 4.21LMVSRR620 pKa = 11.84GVGWDD625 pKa = 3.89GSCPVV630 pKa = 3.74
MM1 pKa = 7.74EE2 pKa = 4.98KK3 pKa = 9.65TPKK6 pKa = 9.07EE7 pKa = 3.79QRR9 pKa = 11.84TPRR12 pKa = 11.84VKK14 pKa = 10.37KK15 pKa = 9.36QQPQKK20 pKa = 10.86KK21 pKa = 8.83PAGQQQPRR29 pKa = 11.84KK30 pKa = 8.66PAQQKK35 pKa = 9.51RR36 pKa = 11.84RR37 pKa = 11.84SPMPQKK43 pKa = 9.18THH45 pKa = 6.97DD46 pKa = 3.67GEE48 pKa = 4.8IYY50 pKa = 10.41SSSSLVSIIHH60 pKa = 6.6GNQDD64 pKa = 4.01DD65 pKa = 4.42FTTPLAISLNPLVVAEE81 pKa = 4.17EE82 pKa = 4.18AKK84 pKa = 11.04DD85 pKa = 3.71PILNMKK91 pKa = 9.47CMSYY95 pKa = 11.13NCFKK99 pKa = 10.98VLSAKK104 pKa = 10.73LEE106 pKa = 4.18LKK108 pKa = 10.19AVCGPNAAVGTTATVGSWQNVDD130 pKa = 5.05DD131 pKa = 4.19PTGPQTPNTALKK143 pKa = 8.98MDD145 pKa = 4.11RR146 pKa = 11.84AVTVGIGMRR155 pKa = 11.84ACAPIKK161 pKa = 10.62HH162 pKa = 5.99SDD164 pKa = 3.55EE165 pKa = 4.34VKK167 pKa = 10.52VVYY170 pKa = 10.06TEE172 pKa = 3.79ATAAPRR178 pKa = 11.84TWNPGHH184 pKa = 7.12AIVSLIGQTGTVYY197 pKa = 10.6NQEE200 pKa = 3.84QTPVWKK206 pKa = 10.37RR207 pKa = 11.84PLWHH211 pKa = 7.05AFLHH215 pKa = 4.81YY216 pKa = 8.86TYY218 pKa = 10.58HH219 pKa = 5.56FTSPKK224 pKa = 10.12DD225 pKa = 3.39NRR227 pKa = 11.84NRR229 pKa = 11.84GSLAAGDD236 pKa = 5.09LDD238 pKa = 3.84DD239 pKa = 5.1QKK241 pKa = 11.71VSLHH245 pKa = 6.72EE246 pKa = 4.62IPDD249 pKa = 3.68EE250 pKa = 4.14PLVLEE255 pKa = 4.81MPNSPAARR263 pKa = 11.84KK264 pKa = 8.98LICRR268 pKa = 11.84GRR270 pKa = 11.84KK271 pKa = 7.69AQGRR275 pKa = 11.84GRR277 pKa = 11.84AGLWDD282 pKa = 4.16FASGILQGLSGVLPPPWGSLLNAGVALVRR311 pKa = 11.84PKK313 pKa = 10.08TATTRR318 pKa = 11.84EE319 pKa = 3.82GDD321 pKa = 3.87TIQLEE326 pKa = 4.39LFQTLSDD333 pKa = 3.88VAQDD337 pKa = 3.62QPVMGVEE344 pKa = 3.93ARR346 pKa = 11.84TTQMDD351 pKa = 5.76LINGHH356 pKa = 5.25YY357 pKa = 9.1TQMTDD362 pKa = 2.9PGANSVLEE370 pKa = 4.53DD371 pKa = 3.53SDD373 pKa = 4.05GGGGVLPPTGGGIVWHH389 pKa = 7.2DD390 pKa = 4.45ANPDD394 pKa = 3.32DD395 pKa = 4.6WSGDD399 pKa = 3.25EE400 pKa = 3.97QIIVNSYY407 pKa = 7.46VNKK410 pKa = 10.53YY411 pKa = 9.55ADD413 pKa = 3.21NYY415 pKa = 9.48FNEE418 pKa = 4.45VYY420 pKa = 9.87PVYY423 pKa = 10.68SSSDD427 pKa = 3.25VNYY430 pKa = 10.6NITVTKK436 pKa = 10.68GPLAGNNYY444 pKa = 8.69QFYY447 pKa = 10.46HH448 pKa = 7.71AMEE451 pKa = 4.18FQKK454 pKa = 10.16WKK456 pKa = 9.4HH457 pKa = 5.26QNTEE461 pKa = 3.81VTAVWVRR468 pKa = 11.84FQKK471 pKa = 9.98MSNMDD476 pKa = 3.95LQAKK480 pKa = 8.63GYY482 pKa = 10.4QMYY485 pKa = 10.5LPQFIEE491 pKa = 4.13KK492 pKa = 8.38FTNASSTSGEE502 pKa = 4.17EE503 pKa = 3.99KK504 pKa = 10.44FVMLAKK510 pKa = 10.73DD511 pKa = 3.5GGRR514 pKa = 11.84VQLQSKK520 pKa = 10.12SSPTVFTRR528 pKa = 11.84FLNMNSSTPWSALFLGEE545 pKa = 4.06SRR547 pKa = 11.84SIKK550 pKa = 9.84IYY552 pKa = 10.56RR553 pKa = 11.84DD554 pKa = 3.59GTSLAAGNWNPCQDD568 pKa = 3.78VLPGNNLSNLQATTAAAGDD587 pKa = 3.98EE588 pKa = 4.43EE589 pKa = 4.37EE590 pKa = 5.56AEE592 pKa = 4.05ADD594 pKa = 5.13KK595 pKa = 11.4EE596 pKa = 4.13DD597 pKa = 3.74LLDD600 pKa = 4.95RR601 pKa = 11.84IRR603 pKa = 11.84YY604 pKa = 8.77LEE606 pKa = 4.45RR607 pKa = 11.84EE608 pKa = 4.37LEE610 pKa = 4.2SAEE613 pKa = 4.14TEE615 pKa = 4.21LMVSRR620 pKa = 11.84GVGWDD625 pKa = 3.89GSCPVV630 pKa = 3.74
Molecular weight: 69.4 kDa
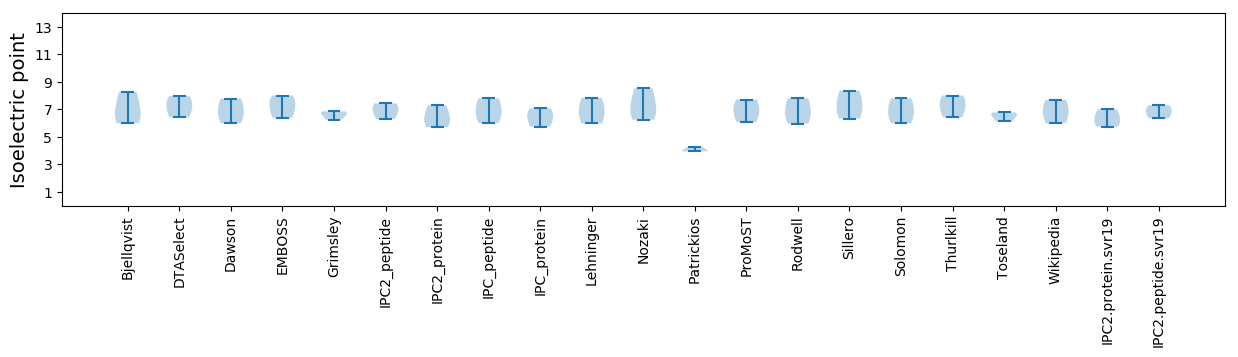
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KNQ2|A0A1L3KNQ2_9VIRU RNA-dependent RNA polymerase OS=Beihai astro-like virus OX=1922353 PE=4 SV=1
MM1 pKa = 8.11SGMLLKK7 pKa = 10.42VLCSVMLLSLSMKK20 pKa = 9.23TDD22 pKa = 4.63AILVVGTCGQVNLPPVATAATAPLMHH48 pKa = 7.16LCTTTNCSVNCDD60 pKa = 3.45VYY62 pKa = 11.25RR63 pKa = 11.84DD64 pKa = 3.74ATRR67 pKa = 11.84YY68 pKa = 8.24PAEE71 pKa = 4.15EE72 pKa = 4.27NEE74 pKa = 4.15LEE76 pKa = 4.07SRR78 pKa = 11.84RR79 pKa = 11.84VRR81 pKa = 11.84RR82 pKa = 11.84HH83 pKa = 5.26CLMGFRR89 pKa = 11.84MNNTYY94 pKa = 10.91GEE96 pKa = 4.48YY97 pKa = 11.06GQFTPPGLSAMTTYY111 pKa = 11.25GNRR114 pKa = 11.84FGLIAPLNQTKK125 pKa = 8.84WEE127 pKa = 3.96NMIIALGGLHH137 pKa = 6.34NCPRR141 pKa = 11.84DD142 pKa = 3.28PTTAMNAAPTPEE154 pKa = 4.06EE155 pKa = 3.84TSAYY159 pKa = 10.36DD160 pKa = 3.64AFHH163 pKa = 6.69KK164 pKa = 9.88TLDD167 pKa = 4.84AIWAHH172 pKa = 5.32YY173 pKa = 10.67NRR175 pKa = 11.84FFSEE179 pKa = 3.92EE180 pKa = 3.72ARR182 pKa = 11.84AKK184 pKa = 10.31AHH186 pKa = 5.04QQRR189 pKa = 11.84LYY191 pKa = 10.4PEE193 pKa = 4.38RR194 pKa = 11.84LEE196 pKa = 4.19HH197 pKa = 6.31LWYY200 pKa = 9.4VQLMMVLSAWWQTLVDD216 pKa = 3.42WHH218 pKa = 6.56IVSAAVMSTFVLTKK232 pKa = 9.93IYY234 pKa = 9.97GQSTTTPWYY243 pKa = 10.24RR244 pKa = 11.84IIALIPAMFTKK255 pKa = 10.62GGLGLALLFTPFVNDD270 pKa = 3.28FNGVLLPMLYY280 pKa = 10.61LLSHH284 pKa = 6.73YY285 pKa = 10.27IYY287 pKa = 10.08LTGGCILIAITLNVGYY303 pKa = 10.98LMGRR307 pKa = 11.84WIVPEE312 pKa = 4.15EE313 pKa = 4.16KK314 pKa = 10.14EE315 pKa = 4.22QDD317 pKa = 3.57GATTNVDD324 pKa = 4.12FMVTTVHH331 pKa = 6.26TARR334 pKa = 11.84SLALVIVACVCGLLATLTPGFPFVLTVMVAISVALILPDD373 pKa = 4.27PPHH376 pKa = 6.48EE377 pKa = 4.44TTAHH381 pKa = 4.86VMVGNGPRR389 pKa = 11.84ARR391 pKa = 11.84RR392 pKa = 11.84VPVKK396 pKa = 10.74VYY398 pKa = 7.3TQPGSRR404 pKa = 11.84NNRR407 pKa = 11.84FWYY410 pKa = 9.76QLQANKK416 pKa = 9.95RR417 pKa = 11.84VQRR420 pKa = 11.84LPEE423 pKa = 3.91TAFGATLTVSQGDD436 pKa = 4.11AISTGFICRR445 pKa = 11.84GMVFTIAHH453 pKa = 5.39GTKK456 pKa = 8.47PTNEE460 pKa = 3.67GMEE463 pKa = 4.03VKK465 pKa = 10.39GPNKK469 pKa = 9.98IVFKK473 pKa = 10.39PGAYY477 pKa = 9.11VGKK480 pKa = 10.19IKK482 pKa = 10.7LSEE485 pKa = 4.25DD486 pKa = 3.17QAVYY490 pKa = 10.7AFKK493 pKa = 11.01LPTEE497 pKa = 4.34ASRR500 pKa = 11.84LKK502 pKa = 10.11SLPVATEE509 pKa = 3.86IKK511 pKa = 10.05EE512 pKa = 4.13GWHH515 pKa = 4.19TVIYY519 pKa = 10.46RR520 pKa = 11.84NEE522 pKa = 3.93DD523 pKa = 3.59CACAGEE529 pKa = 4.23MFYY532 pKa = 11.5GNWEE536 pKa = 4.1DD537 pKa = 4.25DD538 pKa = 4.05EE539 pKa = 5.54LRR541 pKa = 11.84GSYY544 pKa = 10.01TNARR548 pKa = 11.84GYY550 pKa = 10.43SGSPIVNSDD559 pKa = 3.22GKK561 pKa = 9.15ITAVHH566 pKa = 6.4CGGIGEE572 pKa = 4.52VGVSYY577 pKa = 10.94RR578 pKa = 11.84FTAQMIEE585 pKa = 3.95NMTKK589 pKa = 8.98TDD591 pKa = 3.49KK592 pKa = 10.65PKK594 pKa = 9.42KK595 pKa = 8.63TEE597 pKa = 4.0EE598 pKa = 4.03QVEE601 pKa = 4.23DD602 pKa = 3.55QALNPLKK609 pKa = 10.37EE610 pKa = 3.89LALALEE616 pKa = 4.41RR617 pKa = 11.84MTAQFLRR624 pKa = 11.84LEE626 pKa = 4.21AQVRR630 pKa = 11.84DD631 pKa = 3.79NEE633 pKa = 4.33AKK635 pKa = 8.85NTEE638 pKa = 4.06RR639 pKa = 11.84LDD641 pKa = 3.68EE642 pKa = 4.26VEE644 pKa = 5.11NMVNQKK650 pKa = 10.53KK651 pKa = 8.04MFTDD655 pKa = 3.86DD656 pKa = 3.35EE657 pKa = 4.71YY658 pKa = 11.27EE659 pKa = 4.16YY660 pKa = 10.38LTGRR664 pKa = 11.84RR665 pKa = 11.84GMSRR669 pKa = 11.84DD670 pKa = 3.14KK671 pKa = 10.41MRR673 pKa = 11.84EE674 pKa = 3.37IAKK677 pKa = 10.09RR678 pKa = 11.84RR679 pKa = 11.84IAEE682 pKa = 3.87RR683 pKa = 11.84DD684 pKa = 3.17YY685 pKa = 11.8DD686 pKa = 4.06EE687 pKa = 6.44QNDD690 pKa = 3.86QAKK693 pKa = 9.91KK694 pKa = 10.3KK695 pKa = 9.06PDD697 pKa = 3.43VVVASPGALPAIQPTPYY714 pKa = 9.32PAYY717 pKa = 9.62PVVTGKK723 pKa = 10.52APKK726 pKa = 10.1GKK728 pKa = 9.63RR729 pKa = 11.84LHH731 pKa = 5.13EE732 pKa = 4.38WKK734 pKa = 10.52VGMVYY739 pKa = 10.25RR740 pKa = 11.84PNTRR744 pKa = 11.84DD745 pKa = 3.06VHH747 pKa = 7.96VGEE750 pKa = 4.58KK751 pKa = 10.33CCNEE755 pKa = 3.84VVAGYY760 pKa = 10.24EE761 pKa = 4.16NEE763 pKa = 4.47PDD765 pKa = 3.56TQTSHH770 pKa = 7.07IVLTLCTVKK779 pKa = 10.68LVGAGDD785 pKa = 3.65VAYY788 pKa = 10.56CPACEE793 pKa = 3.95PDD795 pKa = 3.22QLRR798 pKa = 11.84FEE800 pKa = 4.61NQRR803 pKa = 11.84GHH805 pKa = 6.89GPLKK809 pKa = 9.06QTKK812 pKa = 8.94CGQKK816 pKa = 6.5TTSMPGHH823 pKa = 6.09VMHH826 pKa = 7.15CKK828 pKa = 8.95ATPKK832 pKa = 9.62CTEE835 pKa = 4.11SDD837 pKa = 4.2CINAPICTDD846 pKa = 3.33HH847 pKa = 6.95CPHH850 pKa = 6.31YY851 pKa = 10.49NCEE854 pKa = 3.85KK855 pKa = 10.25VRR857 pKa = 11.84RR858 pKa = 11.84EE859 pKa = 3.85DD860 pKa = 4.95GIATEE865 pKa = 5.09CPGAKK870 pKa = 9.65CKK872 pKa = 10.49NDD874 pKa = 3.25YY875 pKa = 10.25CKK877 pKa = 10.78KK878 pKa = 10.51KK879 pKa = 10.85GFLKK883 pKa = 10.93GG884 pKa = 3.29
MM1 pKa = 8.11SGMLLKK7 pKa = 10.42VLCSVMLLSLSMKK20 pKa = 9.23TDD22 pKa = 4.63AILVVGTCGQVNLPPVATAATAPLMHH48 pKa = 7.16LCTTTNCSVNCDD60 pKa = 3.45VYY62 pKa = 11.25RR63 pKa = 11.84DD64 pKa = 3.74ATRR67 pKa = 11.84YY68 pKa = 8.24PAEE71 pKa = 4.15EE72 pKa = 4.27NEE74 pKa = 4.15LEE76 pKa = 4.07SRR78 pKa = 11.84RR79 pKa = 11.84VRR81 pKa = 11.84RR82 pKa = 11.84HH83 pKa = 5.26CLMGFRR89 pKa = 11.84MNNTYY94 pKa = 10.91GEE96 pKa = 4.48YY97 pKa = 11.06GQFTPPGLSAMTTYY111 pKa = 11.25GNRR114 pKa = 11.84FGLIAPLNQTKK125 pKa = 8.84WEE127 pKa = 3.96NMIIALGGLHH137 pKa = 6.34NCPRR141 pKa = 11.84DD142 pKa = 3.28PTTAMNAAPTPEE154 pKa = 4.06EE155 pKa = 3.84TSAYY159 pKa = 10.36DD160 pKa = 3.64AFHH163 pKa = 6.69KK164 pKa = 9.88TLDD167 pKa = 4.84AIWAHH172 pKa = 5.32YY173 pKa = 10.67NRR175 pKa = 11.84FFSEE179 pKa = 3.92EE180 pKa = 3.72ARR182 pKa = 11.84AKK184 pKa = 10.31AHH186 pKa = 5.04QQRR189 pKa = 11.84LYY191 pKa = 10.4PEE193 pKa = 4.38RR194 pKa = 11.84LEE196 pKa = 4.19HH197 pKa = 6.31LWYY200 pKa = 9.4VQLMMVLSAWWQTLVDD216 pKa = 3.42WHH218 pKa = 6.56IVSAAVMSTFVLTKK232 pKa = 9.93IYY234 pKa = 9.97GQSTTTPWYY243 pKa = 10.24RR244 pKa = 11.84IIALIPAMFTKK255 pKa = 10.62GGLGLALLFTPFVNDD270 pKa = 3.28FNGVLLPMLYY280 pKa = 10.61LLSHH284 pKa = 6.73YY285 pKa = 10.27IYY287 pKa = 10.08LTGGCILIAITLNVGYY303 pKa = 10.98LMGRR307 pKa = 11.84WIVPEE312 pKa = 4.15EE313 pKa = 4.16KK314 pKa = 10.14EE315 pKa = 4.22QDD317 pKa = 3.57GATTNVDD324 pKa = 4.12FMVTTVHH331 pKa = 6.26TARR334 pKa = 11.84SLALVIVACVCGLLATLTPGFPFVLTVMVAISVALILPDD373 pKa = 4.27PPHH376 pKa = 6.48EE377 pKa = 4.44TTAHH381 pKa = 4.86VMVGNGPRR389 pKa = 11.84ARR391 pKa = 11.84RR392 pKa = 11.84VPVKK396 pKa = 10.74VYY398 pKa = 7.3TQPGSRR404 pKa = 11.84NNRR407 pKa = 11.84FWYY410 pKa = 9.76QLQANKK416 pKa = 9.95RR417 pKa = 11.84VQRR420 pKa = 11.84LPEE423 pKa = 3.91TAFGATLTVSQGDD436 pKa = 4.11AISTGFICRR445 pKa = 11.84GMVFTIAHH453 pKa = 5.39GTKK456 pKa = 8.47PTNEE460 pKa = 3.67GMEE463 pKa = 4.03VKK465 pKa = 10.39GPNKK469 pKa = 9.98IVFKK473 pKa = 10.39PGAYY477 pKa = 9.11VGKK480 pKa = 10.19IKK482 pKa = 10.7LSEE485 pKa = 4.25DD486 pKa = 3.17QAVYY490 pKa = 10.7AFKK493 pKa = 11.01LPTEE497 pKa = 4.34ASRR500 pKa = 11.84LKK502 pKa = 10.11SLPVATEE509 pKa = 3.86IKK511 pKa = 10.05EE512 pKa = 4.13GWHH515 pKa = 4.19TVIYY519 pKa = 10.46RR520 pKa = 11.84NEE522 pKa = 3.93DD523 pKa = 3.59CACAGEE529 pKa = 4.23MFYY532 pKa = 11.5GNWEE536 pKa = 4.1DD537 pKa = 4.25DD538 pKa = 4.05EE539 pKa = 5.54LRR541 pKa = 11.84GSYY544 pKa = 10.01TNARR548 pKa = 11.84GYY550 pKa = 10.43SGSPIVNSDD559 pKa = 3.22GKK561 pKa = 9.15ITAVHH566 pKa = 6.4CGGIGEE572 pKa = 4.52VGVSYY577 pKa = 10.94RR578 pKa = 11.84FTAQMIEE585 pKa = 3.95NMTKK589 pKa = 8.98TDD591 pKa = 3.49KK592 pKa = 10.65PKK594 pKa = 9.42KK595 pKa = 8.63TEE597 pKa = 4.0EE598 pKa = 4.03QVEE601 pKa = 4.23DD602 pKa = 3.55QALNPLKK609 pKa = 10.37EE610 pKa = 3.89LALALEE616 pKa = 4.41RR617 pKa = 11.84MTAQFLRR624 pKa = 11.84LEE626 pKa = 4.21AQVRR630 pKa = 11.84DD631 pKa = 3.79NEE633 pKa = 4.33AKK635 pKa = 8.85NTEE638 pKa = 4.06RR639 pKa = 11.84LDD641 pKa = 3.68EE642 pKa = 4.26VEE644 pKa = 5.11NMVNQKK650 pKa = 10.53KK651 pKa = 8.04MFTDD655 pKa = 3.86DD656 pKa = 3.35EE657 pKa = 4.71YY658 pKa = 11.27EE659 pKa = 4.16YY660 pKa = 10.38LTGRR664 pKa = 11.84RR665 pKa = 11.84GMSRR669 pKa = 11.84DD670 pKa = 3.14KK671 pKa = 10.41MRR673 pKa = 11.84EE674 pKa = 3.37IAKK677 pKa = 10.09RR678 pKa = 11.84RR679 pKa = 11.84IAEE682 pKa = 3.87RR683 pKa = 11.84DD684 pKa = 3.17YY685 pKa = 11.8DD686 pKa = 4.06EE687 pKa = 6.44QNDD690 pKa = 3.86QAKK693 pKa = 9.91KK694 pKa = 10.3KK695 pKa = 9.06PDD697 pKa = 3.43VVVASPGALPAIQPTPYY714 pKa = 9.32PAYY717 pKa = 9.62PVVTGKK723 pKa = 10.52APKK726 pKa = 10.1GKK728 pKa = 9.63RR729 pKa = 11.84LHH731 pKa = 5.13EE732 pKa = 4.38WKK734 pKa = 10.52VGMVYY739 pKa = 10.25RR740 pKa = 11.84PNTRR744 pKa = 11.84DD745 pKa = 3.06VHH747 pKa = 7.96VGEE750 pKa = 4.58KK751 pKa = 10.33CCNEE755 pKa = 3.84VVAGYY760 pKa = 10.24EE761 pKa = 4.16NEE763 pKa = 4.47PDD765 pKa = 3.56TQTSHH770 pKa = 7.07IVLTLCTVKK779 pKa = 10.68LVGAGDD785 pKa = 3.65VAYY788 pKa = 10.56CPACEE793 pKa = 3.95PDD795 pKa = 3.22QLRR798 pKa = 11.84FEE800 pKa = 4.61NQRR803 pKa = 11.84GHH805 pKa = 6.89GPLKK809 pKa = 9.06QTKK812 pKa = 8.94CGQKK816 pKa = 6.5TTSMPGHH823 pKa = 6.09VMHH826 pKa = 7.15CKK828 pKa = 8.95ATPKK832 pKa = 9.62CTEE835 pKa = 4.11SDD837 pKa = 4.2CINAPICTDD846 pKa = 3.33HH847 pKa = 6.95CPHH850 pKa = 6.31YY851 pKa = 10.49NCEE854 pKa = 3.85KK855 pKa = 10.25VRR857 pKa = 11.84RR858 pKa = 11.84EE859 pKa = 3.85DD860 pKa = 4.95GIATEE865 pKa = 5.09CPGAKK870 pKa = 9.65CKK872 pKa = 10.49NDD874 pKa = 3.25YY875 pKa = 10.25CKK877 pKa = 10.78KK878 pKa = 10.51KK879 pKa = 10.85GFLKK883 pKa = 10.93GG884 pKa = 3.29
Molecular weight: 98.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1905 |
391 |
884 |
635.0 |
70.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.717 ± 0.449 | 2.362 ± 0.609 |
4.829 ± 0.467 | 6.194 ± 0.416 |
2.835 ± 0.131 | 7.192 ± 0.333 |
2.415 ± 0.226 | 3.937 ± 0.201 |
5.722 ± 0.075 | 7.979 ± 0.159 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.255 ± 0.26 | 4.829 ± 0.253 |
5.932 ± 0.374 | 4.094 ± 0.785 |
5.249 ± 0.406 | 4.987 ± 0.945 |
7.717 ± 0.407 | 7.139 ± 0.341 |
1.68 ± 0.2 | 3.937 ± 0.53 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
