
Jiangella asiatica
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Jiangellales; Jiangellaceae; Jiangella
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
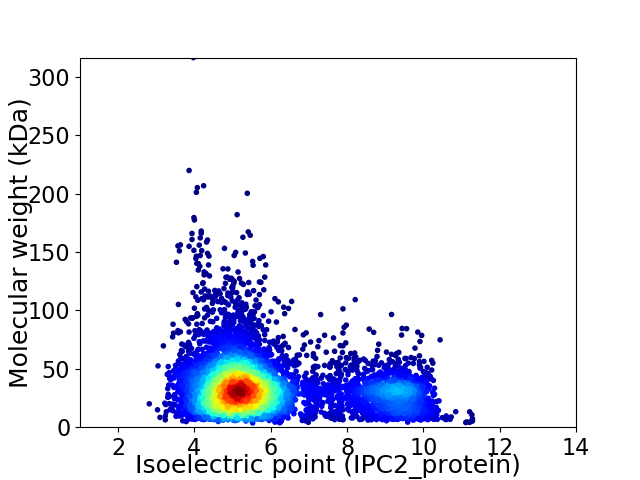
Virtual 2D-PAGE plot for 6206 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R5DLG4|A0A4R5DLG4_9ACTN VOC family protein OS=Jiangella asiatica OX=2530372 GN=E1269_09470 PE=4 SV=1
MM1 pKa = 7.63AARR4 pKa = 11.84AGAAALGIAALVSACGGGGEE24 pKa = 4.6EE25 pKa = 5.15VDD27 pKa = 3.71VDD29 pKa = 4.5DD30 pKa = 6.27GNGTSGADD38 pKa = 3.22QGGGDD43 pKa = 3.95GDD45 pKa = 4.17APAGTLVAAISAQPDD60 pKa = 3.11QFDD63 pKa = 3.48PHH65 pKa = 6.08KK66 pKa = 8.27TQAYY70 pKa = 10.15ASFQVLEE77 pKa = 4.21NVYY80 pKa = 9.04DD81 pKa = 3.9TLVVPDD87 pKa = 4.79AEE89 pKa = 5.69GEE91 pKa = 4.27FQPSLATEE99 pKa = 4.23WTTSDD104 pKa = 5.56DD105 pKa = 3.52ALTWTFTLRR114 pKa = 11.84EE115 pKa = 4.25GVTFHH120 pKa = 7.47DD121 pKa = 5.21GSAFDD126 pKa = 3.98SADD129 pKa = 2.91VVYY132 pKa = 10.33SYY134 pKa = 11.3RR135 pKa = 11.84RR136 pKa = 11.84IIDD139 pKa = 3.47EE140 pKa = 4.05QLANAYY146 pKa = 9.95RR147 pKa = 11.84FASVANIDD155 pKa = 3.9APDD158 pKa = 4.13PQTVVITLSTPTPALLDD175 pKa = 4.02NIGGFKK181 pKa = 10.51GMAIIPEE188 pKa = 4.43GAGQQTDD195 pKa = 3.56LATTAVGTGPFTLEE209 pKa = 3.52EE210 pKa = 4.11TGAGGVTLARR220 pKa = 11.84YY221 pKa = 9.7DD222 pKa = 4.83DD223 pKa = 4.09YY224 pKa = 11.26WGDD227 pKa = 3.44PATVEE232 pKa = 4.25GVEE235 pKa = 4.06FRR237 pKa = 11.84YY238 pKa = 10.49VSEE241 pKa = 3.99PAAALTALRR250 pKa = 11.84SGDD253 pKa = 3.31IHH255 pKa = 5.96WTDD258 pKa = 3.37NVPPQDD264 pKa = 4.07IEE266 pKa = 4.29GLQDD270 pKa = 3.67DD271 pKa = 4.94EE272 pKa = 4.44EE273 pKa = 5.3VEE275 pKa = 4.57LGVTPSVDD283 pKa = 3.06YY284 pKa = 9.78WYY286 pKa = 11.06LAMNQTRR293 pKa = 11.84PPFDD297 pKa = 3.14NRR299 pKa = 11.84DD300 pKa = 3.3FRR302 pKa = 11.84RR303 pKa = 11.84GVAYY307 pKa = 10.06GIDD310 pKa = 3.39RR311 pKa = 11.84EE312 pKa = 4.53EE313 pKa = 3.89VTEE316 pKa = 3.92AARR319 pKa = 11.84FGAATTNQTAIPEE332 pKa = 4.05QSLWYY337 pKa = 10.06HH338 pKa = 6.62EE339 pKa = 4.32YY340 pKa = 11.22APFEE344 pKa = 4.53HH345 pKa = 7.51DD346 pKa = 3.58PDD348 pKa = 3.56QAADD352 pKa = 4.56LIEE355 pKa = 4.16QTAAATPQPMGLMVTDD371 pKa = 5.18EE372 pKa = 4.32YY373 pKa = 10.95PEE375 pKa = 4.32TVQAAQVIQAQLAEE389 pKa = 4.25VGVEE393 pKa = 3.59IGIEE397 pKa = 3.93QEE399 pKa = 5.82AFATWLDD406 pKa = 3.56RR407 pKa = 11.84QAAGDD412 pKa = 3.79YY413 pKa = 11.03DD414 pKa = 4.33AFLLGWLGNLDD425 pKa = 3.53PFGFYY430 pKa = 10.0HH431 pKa = 6.36AQHH434 pKa = 5.75VCEE437 pKa = 4.48GTSNFQGYY445 pKa = 9.78CNPEE449 pKa = 3.53VDD451 pKa = 3.67TLLNQAAAEE460 pKa = 4.12TDD462 pKa = 3.39QEE464 pKa = 4.16ARR466 pKa = 11.84KK467 pKa = 9.78ALYY470 pKa = 10.04DD471 pKa = 3.48QAAEE475 pKa = 4.73LIVDD479 pKa = 3.99DD480 pKa = 4.67VSYY483 pKa = 10.96LYY485 pKa = 10.58LYY487 pKa = 10.65NPDD490 pKa = 4.08VVQAWAPGLEE500 pKa = 4.5GYY502 pKa = 9.03EE503 pKa = 3.9IRR505 pKa = 11.84PDD507 pKa = 3.15KK508 pKa = 10.89AINFEE513 pKa = 4.46TVRR516 pKa = 11.84LPEE519 pKa = 3.86
MM1 pKa = 7.63AARR4 pKa = 11.84AGAAALGIAALVSACGGGGEE24 pKa = 4.6EE25 pKa = 5.15VDD27 pKa = 3.71VDD29 pKa = 4.5DD30 pKa = 6.27GNGTSGADD38 pKa = 3.22QGGGDD43 pKa = 3.95GDD45 pKa = 4.17APAGTLVAAISAQPDD60 pKa = 3.11QFDD63 pKa = 3.48PHH65 pKa = 6.08KK66 pKa = 8.27TQAYY70 pKa = 10.15ASFQVLEE77 pKa = 4.21NVYY80 pKa = 9.04DD81 pKa = 3.9TLVVPDD87 pKa = 4.79AEE89 pKa = 5.69GEE91 pKa = 4.27FQPSLATEE99 pKa = 4.23WTTSDD104 pKa = 5.56DD105 pKa = 3.52ALTWTFTLRR114 pKa = 11.84EE115 pKa = 4.25GVTFHH120 pKa = 7.47DD121 pKa = 5.21GSAFDD126 pKa = 3.98SADD129 pKa = 2.91VVYY132 pKa = 10.33SYY134 pKa = 11.3RR135 pKa = 11.84RR136 pKa = 11.84IIDD139 pKa = 3.47EE140 pKa = 4.05QLANAYY146 pKa = 9.95RR147 pKa = 11.84FASVANIDD155 pKa = 3.9APDD158 pKa = 4.13PQTVVITLSTPTPALLDD175 pKa = 4.02NIGGFKK181 pKa = 10.51GMAIIPEE188 pKa = 4.43GAGQQTDD195 pKa = 3.56LATTAVGTGPFTLEE209 pKa = 3.52EE210 pKa = 4.11TGAGGVTLARR220 pKa = 11.84YY221 pKa = 9.7DD222 pKa = 4.83DD223 pKa = 4.09YY224 pKa = 11.26WGDD227 pKa = 3.44PATVEE232 pKa = 4.25GVEE235 pKa = 4.06FRR237 pKa = 11.84YY238 pKa = 10.49VSEE241 pKa = 3.99PAAALTALRR250 pKa = 11.84SGDD253 pKa = 3.31IHH255 pKa = 5.96WTDD258 pKa = 3.37NVPPQDD264 pKa = 4.07IEE266 pKa = 4.29GLQDD270 pKa = 3.67DD271 pKa = 4.94EE272 pKa = 4.44EE273 pKa = 5.3VEE275 pKa = 4.57LGVTPSVDD283 pKa = 3.06YY284 pKa = 9.78WYY286 pKa = 11.06LAMNQTRR293 pKa = 11.84PPFDD297 pKa = 3.14NRR299 pKa = 11.84DD300 pKa = 3.3FRR302 pKa = 11.84RR303 pKa = 11.84GVAYY307 pKa = 10.06GIDD310 pKa = 3.39RR311 pKa = 11.84EE312 pKa = 4.53EE313 pKa = 3.89VTEE316 pKa = 3.92AARR319 pKa = 11.84FGAATTNQTAIPEE332 pKa = 4.05QSLWYY337 pKa = 10.06HH338 pKa = 6.62EE339 pKa = 4.32YY340 pKa = 11.22APFEE344 pKa = 4.53HH345 pKa = 7.51DD346 pKa = 3.58PDD348 pKa = 3.56QAADD352 pKa = 4.56LIEE355 pKa = 4.16QTAAATPQPMGLMVTDD371 pKa = 5.18EE372 pKa = 4.32YY373 pKa = 10.95PEE375 pKa = 4.32TVQAAQVIQAQLAEE389 pKa = 4.25VGVEE393 pKa = 3.59IGIEE397 pKa = 3.93QEE399 pKa = 5.82AFATWLDD406 pKa = 3.56RR407 pKa = 11.84QAAGDD412 pKa = 3.79YY413 pKa = 11.03DD414 pKa = 4.33AFLLGWLGNLDD425 pKa = 3.53PFGFYY430 pKa = 10.0HH431 pKa = 6.36AQHH434 pKa = 5.75VCEE437 pKa = 4.48GTSNFQGYY445 pKa = 9.78CNPEE449 pKa = 3.53VDD451 pKa = 3.67TLLNQAAAEE460 pKa = 4.12TDD462 pKa = 3.39QEE464 pKa = 4.16ARR466 pKa = 11.84KK467 pKa = 9.78ALYY470 pKa = 10.04DD471 pKa = 3.48QAAEE475 pKa = 4.73LIVDD479 pKa = 3.99DD480 pKa = 4.67VSYY483 pKa = 10.96LYY485 pKa = 10.58LYY487 pKa = 10.65NPDD490 pKa = 4.08VVQAWAPGLEE500 pKa = 4.5GYY502 pKa = 9.03EE503 pKa = 3.9IRR505 pKa = 11.84PDD507 pKa = 3.15KK508 pKa = 10.89AINFEE513 pKa = 4.46TVRR516 pKa = 11.84LPEE519 pKa = 3.86
Molecular weight: 55.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R5CFL7|A0A4R5CFL7_9ACTN Uncharacterized protein OS=Jiangella asiatica OX=2530372 GN=E1269_28210 PE=4 SV=1
PP1 pKa = 7.17HH2 pKa = 7.43RR3 pKa = 11.84NLRR6 pKa = 11.84PGPRR10 pKa = 11.84PRR12 pKa = 11.84PRR14 pKa = 11.84HH15 pKa = 5.02PHH17 pKa = 4.75HH18 pKa = 6.21QRR20 pKa = 11.84PHH22 pKa = 6.16RR23 pKa = 11.84RR24 pKa = 11.84TPTRR28 pKa = 11.84ARR30 pKa = 11.84PGPHH34 pKa = 5.2THH36 pKa = 6.95LPAHH40 pKa = 7.07RR41 pKa = 11.84PPPGPTRR48 pKa = 11.84KK49 pKa = 9.79
PP1 pKa = 7.17HH2 pKa = 7.43RR3 pKa = 11.84NLRR6 pKa = 11.84PGPRR10 pKa = 11.84PRR12 pKa = 11.84PRR14 pKa = 11.84HH15 pKa = 5.02PHH17 pKa = 4.75HH18 pKa = 6.21QRR20 pKa = 11.84PHH22 pKa = 6.16RR23 pKa = 11.84RR24 pKa = 11.84TPTRR28 pKa = 11.84ARR30 pKa = 11.84PGPHH34 pKa = 5.2THH36 pKa = 6.95LPAHH40 pKa = 7.07RR41 pKa = 11.84PPPGPTRR48 pKa = 11.84KK49 pKa = 9.79
Molecular weight: 5.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2091672 |
33 |
3131 |
337.0 |
36.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.357 ± 0.041 | 0.682 ± 0.008 |
6.754 ± 0.03 | 5.571 ± 0.025 |
2.826 ± 0.019 | 9.375 ± 0.028 |
2.179 ± 0.014 | 3.459 ± 0.02 |
1.317 ± 0.015 | 10.035 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.757 ± 0.011 | 1.774 ± 0.015 |
5.928 ± 0.024 | 2.614 ± 0.013 |
7.955 ± 0.041 | 5.132 ± 0.018 |
6.115 ± 0.028 | 9.433 ± 0.031 |
1.616 ± 0.014 | 2.121 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
