
Crimson clover cryptic virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Betapartitivirus
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
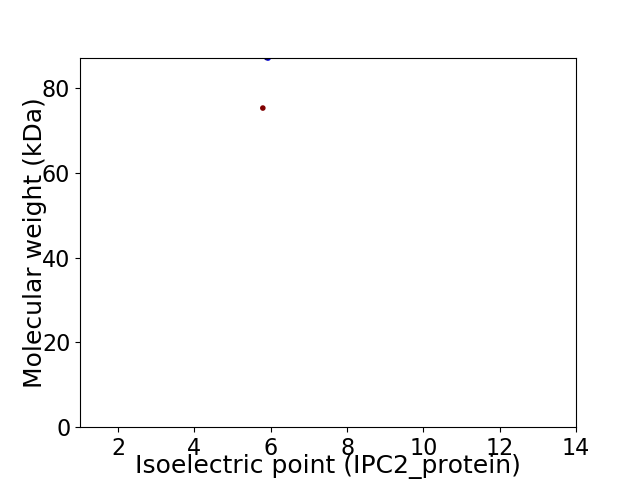
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M9VTV7|M9VTV7_9VIRU RNA-dependent RNA polymerase OS=Crimson clover cryptic virus 2 OX=1323528 PE=4 SV=1
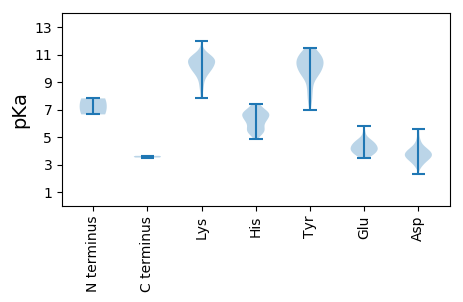
MM1 pKa = 6.65TTNSSNARR9 pKa = 11.84LNKK12 pKa = 10.01LKK14 pKa = 10.96NNTVPLPFNATEE26 pKa = 4.25VPEE29 pKa = 4.37PATDD33 pKa = 4.05LLTSMKK39 pKa = 9.92EE40 pKa = 3.71NTSAHH45 pKa = 6.22SATANQWTIDD55 pKa = 4.26LFPNMIPVLIHH66 pKa = 5.69VMSAASHH73 pKa = 6.21HH74 pKa = 6.58ANAINFRR81 pKa = 11.84EE82 pKa = 3.99HH83 pKa = 6.8SKK85 pKa = 9.98TSAATVAMYY94 pKa = 10.8HH95 pKa = 5.44MAVVYY100 pKa = 9.69GYY102 pKa = 11.15FLLNDD107 pKa = 3.75LNIRR111 pKa = 11.84LNPSAHH117 pKa = 5.41ARR119 pKa = 11.84SWAEE123 pKa = 3.72ASWKK127 pKa = 10.63SEE129 pKa = 3.62FVEE132 pKa = 4.31FLYY135 pKa = 10.85GLPVPEE141 pKa = 4.81FLAPILSEE149 pKa = 4.01YY150 pKa = 9.56QHH152 pKa = 6.52FVTDD156 pKa = 3.54KK157 pKa = 9.32TPNVHH162 pKa = 6.1FTPSAAGFDD171 pKa = 3.42HH172 pKa = 7.29DD173 pKa = 4.07QFFGRR178 pKa = 11.84VFPLNMFAALHH189 pKa = 6.88DD190 pKa = 4.23STATLPSSSTYY201 pKa = 10.92AQVLQDD207 pKa = 3.98LFSRR211 pKa = 11.84PLYY214 pKa = 10.18TITDD218 pKa = 3.97PAFTCYY224 pKa = 10.54LPDD227 pKa = 3.95LLGITLTAGEE237 pKa = 4.52NVTTLNYY244 pKa = 9.87INSKK248 pKa = 9.92LYY250 pKa = 10.61QVFTTIFNPVLFRR263 pKa = 11.84DD264 pKa = 3.71HH265 pKa = 5.86QRR267 pKa = 11.84RR268 pKa = 11.84SSLAALSFKK277 pKa = 11.0SPTYY281 pKa = 10.11PNPNINAYY289 pKa = 10.03DD290 pKa = 4.04LLFSASVANLRR301 pKa = 11.84EE302 pKa = 4.15IKK304 pKa = 10.2IVLQSIYY311 pKa = 10.83KK312 pKa = 10.44LFDD315 pKa = 3.01GRR317 pKa = 11.84VACKK321 pKa = 8.81HH322 pKa = 5.5TLSQFICEE330 pKa = 4.3STSPSITKK338 pKa = 9.82HH339 pKa = 5.7GYY341 pKa = 7.84STFPLPTWSHH351 pKa = 5.78TEE353 pKa = 3.61SDD355 pKa = 3.07AKK357 pKa = 10.62AIRR360 pKa = 11.84FSGVTALVHH369 pKa = 6.22VSEE372 pKa = 4.87EE373 pKa = 3.76DD374 pKa = 3.33RR375 pKa = 11.84AQDD378 pKa = 3.19FCFLQRR384 pKa = 11.84PAEE387 pKa = 4.67AIPHH391 pKa = 5.21VNEE394 pKa = 4.16VPDD397 pKa = 3.26IRR399 pKa = 11.84YY400 pKa = 7.49ATTEE404 pKa = 4.02DD405 pKa = 4.35PEE407 pKa = 4.63TAVPLPANHH416 pKa = 6.28ILVRR420 pKa = 11.84RR421 pKa = 11.84FPFAIRR427 pKa = 11.84LRR429 pKa = 11.84QEE431 pKa = 3.8AANGFPRR438 pKa = 11.84HH439 pKa = 6.74DD440 pKa = 4.16NDD442 pKa = 5.43DD443 pKa = 3.54LTKK446 pKa = 10.73FSDD449 pKa = 3.99HH450 pKa = 5.33VHH452 pKa = 5.51TAPRR456 pKa = 11.84VLILDD461 pKa = 3.7TTGDD465 pKa = 4.26GIISAHH471 pKa = 5.21TTTASGKK478 pKa = 9.58IIEE481 pKa = 4.39SFEE484 pKa = 4.67LDD486 pKa = 3.04GSTIEE491 pKa = 4.75MPNADD496 pKa = 4.58KK497 pKa = 11.17SLGMQNCLFADD508 pKa = 3.42SAIAYY513 pKa = 8.89KK514 pKa = 9.6YY515 pKa = 9.43VRR517 pKa = 11.84PGSFYY522 pKa = 10.78RR523 pKa = 11.84PRR525 pKa = 11.84TQGSVLPPLNRR536 pKa = 11.84APPNSRR542 pKa = 11.84PRR544 pKa = 11.84LPASSMLHH552 pKa = 6.68DD553 pKa = 3.68RR554 pKa = 11.84TKK556 pKa = 11.13VFLPQLNTRR565 pKa = 11.84INEE568 pKa = 4.06AVDD571 pKa = 3.04IRR573 pKa = 11.84TLPGLTMLNNVNCINYY589 pKa = 8.46AQSFIGFKK597 pKa = 10.19TVDD600 pKa = 2.95ATRR603 pKa = 11.84NDD605 pKa = 3.96DD606 pKa = 3.81VLDD609 pKa = 4.29AVPGMPLNNLILWSPYY625 pKa = 9.86TYY627 pKa = 9.79TAYY630 pKa = 10.19EE631 pKa = 4.44DD632 pKa = 3.49EE633 pKa = 4.85TYY635 pKa = 10.56PDD637 pKa = 3.83PVFSNSRR644 pKa = 11.84HH645 pKa = 5.29YY646 pKa = 11.33YY647 pKa = 7.96LTNLRR652 pKa = 11.84TIFGTDD658 pKa = 2.85TNLIKK663 pKa = 10.53AVHH666 pKa = 6.82PYY668 pKa = 9.41EE669 pKa = 4.91AYY671 pKa = 9.61PVSS674 pKa = 3.51
MM1 pKa = 6.65TTNSSNARR9 pKa = 11.84LNKK12 pKa = 10.01LKK14 pKa = 10.96NNTVPLPFNATEE26 pKa = 4.25VPEE29 pKa = 4.37PATDD33 pKa = 4.05LLTSMKK39 pKa = 9.92EE40 pKa = 3.71NTSAHH45 pKa = 6.22SATANQWTIDD55 pKa = 4.26LFPNMIPVLIHH66 pKa = 5.69VMSAASHH73 pKa = 6.21HH74 pKa = 6.58ANAINFRR81 pKa = 11.84EE82 pKa = 3.99HH83 pKa = 6.8SKK85 pKa = 9.98TSAATVAMYY94 pKa = 10.8HH95 pKa = 5.44MAVVYY100 pKa = 9.69GYY102 pKa = 11.15FLLNDD107 pKa = 3.75LNIRR111 pKa = 11.84LNPSAHH117 pKa = 5.41ARR119 pKa = 11.84SWAEE123 pKa = 3.72ASWKK127 pKa = 10.63SEE129 pKa = 3.62FVEE132 pKa = 4.31FLYY135 pKa = 10.85GLPVPEE141 pKa = 4.81FLAPILSEE149 pKa = 4.01YY150 pKa = 9.56QHH152 pKa = 6.52FVTDD156 pKa = 3.54KK157 pKa = 9.32TPNVHH162 pKa = 6.1FTPSAAGFDD171 pKa = 3.42HH172 pKa = 7.29DD173 pKa = 4.07QFFGRR178 pKa = 11.84VFPLNMFAALHH189 pKa = 6.88DD190 pKa = 4.23STATLPSSSTYY201 pKa = 10.92AQVLQDD207 pKa = 3.98LFSRR211 pKa = 11.84PLYY214 pKa = 10.18TITDD218 pKa = 3.97PAFTCYY224 pKa = 10.54LPDD227 pKa = 3.95LLGITLTAGEE237 pKa = 4.52NVTTLNYY244 pKa = 9.87INSKK248 pKa = 9.92LYY250 pKa = 10.61QVFTTIFNPVLFRR263 pKa = 11.84DD264 pKa = 3.71HH265 pKa = 5.86QRR267 pKa = 11.84RR268 pKa = 11.84SSLAALSFKK277 pKa = 11.0SPTYY281 pKa = 10.11PNPNINAYY289 pKa = 10.03DD290 pKa = 4.04LLFSASVANLRR301 pKa = 11.84EE302 pKa = 4.15IKK304 pKa = 10.2IVLQSIYY311 pKa = 10.83KK312 pKa = 10.44LFDD315 pKa = 3.01GRR317 pKa = 11.84VACKK321 pKa = 8.81HH322 pKa = 5.5TLSQFICEE330 pKa = 4.3STSPSITKK338 pKa = 9.82HH339 pKa = 5.7GYY341 pKa = 7.84STFPLPTWSHH351 pKa = 5.78TEE353 pKa = 3.61SDD355 pKa = 3.07AKK357 pKa = 10.62AIRR360 pKa = 11.84FSGVTALVHH369 pKa = 6.22VSEE372 pKa = 4.87EE373 pKa = 3.76DD374 pKa = 3.33RR375 pKa = 11.84AQDD378 pKa = 3.19FCFLQRR384 pKa = 11.84PAEE387 pKa = 4.67AIPHH391 pKa = 5.21VNEE394 pKa = 4.16VPDD397 pKa = 3.26IRR399 pKa = 11.84YY400 pKa = 7.49ATTEE404 pKa = 4.02DD405 pKa = 4.35PEE407 pKa = 4.63TAVPLPANHH416 pKa = 6.28ILVRR420 pKa = 11.84RR421 pKa = 11.84FPFAIRR427 pKa = 11.84LRR429 pKa = 11.84QEE431 pKa = 3.8AANGFPRR438 pKa = 11.84HH439 pKa = 6.74DD440 pKa = 4.16NDD442 pKa = 5.43DD443 pKa = 3.54LTKK446 pKa = 10.73FSDD449 pKa = 3.99HH450 pKa = 5.33VHH452 pKa = 5.51TAPRR456 pKa = 11.84VLILDD461 pKa = 3.7TTGDD465 pKa = 4.26GIISAHH471 pKa = 5.21TTTASGKK478 pKa = 9.58IIEE481 pKa = 4.39SFEE484 pKa = 4.67LDD486 pKa = 3.04GSTIEE491 pKa = 4.75MPNADD496 pKa = 4.58KK497 pKa = 11.17SLGMQNCLFADD508 pKa = 3.42SAIAYY513 pKa = 8.89KK514 pKa = 9.6YY515 pKa = 9.43VRR517 pKa = 11.84PGSFYY522 pKa = 10.78RR523 pKa = 11.84PRR525 pKa = 11.84TQGSVLPPLNRR536 pKa = 11.84APPNSRR542 pKa = 11.84PRR544 pKa = 11.84LPASSMLHH552 pKa = 6.68DD553 pKa = 3.68RR554 pKa = 11.84TKK556 pKa = 11.13VFLPQLNTRR565 pKa = 11.84INEE568 pKa = 4.06AVDD571 pKa = 3.04IRR573 pKa = 11.84TLPGLTMLNNVNCINYY589 pKa = 8.46AQSFIGFKK597 pKa = 10.19TVDD600 pKa = 2.95ATRR603 pKa = 11.84NDD605 pKa = 3.96DD606 pKa = 3.81VLDD609 pKa = 4.29AVPGMPLNNLILWSPYY625 pKa = 9.86TYY627 pKa = 9.79TAYY630 pKa = 10.19EE631 pKa = 4.44DD632 pKa = 3.49EE633 pKa = 4.85TYY635 pKa = 10.56PDD637 pKa = 3.83PVFSNSRR644 pKa = 11.84HH645 pKa = 5.29YY646 pKa = 11.33YY647 pKa = 7.96LTNLRR652 pKa = 11.84TIFGTDD658 pKa = 2.85TNLIKK663 pKa = 10.53AVHH666 pKa = 6.82PYY668 pKa = 9.41EE669 pKa = 4.91AYY671 pKa = 9.61PVSS674 pKa = 3.51
Molecular weight: 75.36 kDa
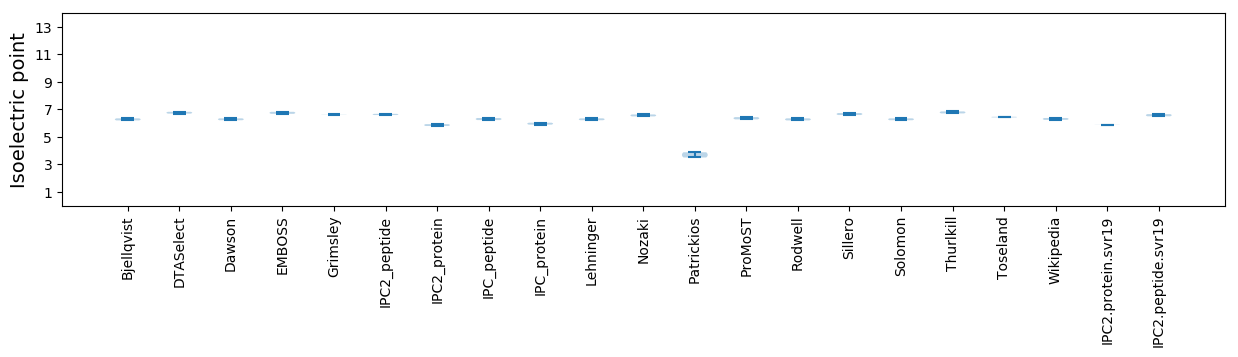
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M9VTV7|M9VTV7_9VIRU RNA-dependent RNA polymerase OS=Crimson clover cryptic virus 2 OX=1323528 PE=4 SV=1
MM1 pKa = 7.81PFNAVRR7 pKa = 11.84NYY9 pKa = 10.52LNEE12 pKa = 3.53RR13 pKa = 11.84MTRR16 pKa = 11.84LKK18 pKa = 10.63QEE20 pKa = 3.51WKK22 pKa = 8.49TYY24 pKa = 10.3QSTDD28 pKa = 2.96HH29 pKa = 7.31DD30 pKa = 5.06PIKK33 pKa = 10.71ILDD36 pKa = 4.3TIQDD40 pKa = 3.59PDD42 pKa = 3.32YY43 pKa = 11.23RR44 pKa = 11.84RR45 pKa = 11.84YY46 pKa = 10.67LDD48 pKa = 3.33NARR51 pKa = 11.84FNSDD55 pKa = 3.17NEE57 pKa = 4.48MKK59 pKa = 10.57HH60 pKa = 6.7LMLNKK65 pKa = 9.91EE66 pKa = 3.89YY67 pKa = 9.61STLVEE72 pKa = 5.36AYY74 pKa = 8.28RR75 pKa = 11.84TDD77 pKa = 3.31NAKK80 pKa = 9.94KK81 pKa = 7.82HH82 pKa = 4.88QVYY85 pKa = 9.84EE86 pKa = 3.83LHH88 pKa = 6.59QPIPNDD94 pKa = 3.58AAPVIQSRR102 pKa = 11.84LPAKK106 pKa = 10.1GIKK109 pKa = 9.39LVPLMYY115 pKa = 10.13HH116 pKa = 6.17YY117 pKa = 11.45GHH119 pKa = 6.32VTHH122 pKa = 7.42DD123 pKa = 3.91PVSLTDD129 pKa = 4.02ANRR132 pKa = 11.84SDD134 pKa = 4.71DD135 pKa = 4.44FEE137 pKa = 4.61TQVVPDD143 pKa = 3.73SDD145 pKa = 3.55APTRR149 pKa = 11.84FGYY152 pKa = 10.0PIDD155 pKa = 3.56VRR157 pKa = 11.84IYY159 pKa = 10.33NLICYY164 pKa = 9.56RR165 pKa = 11.84YY166 pKa = 8.73SKK168 pKa = 10.59YY169 pKa = 11.01LEE171 pKa = 4.51VINAYY176 pKa = 9.52CRR178 pKa = 11.84PIGTVNATFEE188 pKa = 4.58DD189 pKa = 4.7FNKK192 pKa = 10.12EE193 pKa = 4.12QIPSAPIDD201 pKa = 3.67PKK203 pKa = 10.73RR204 pKa = 11.84KK205 pKa = 9.58EE206 pKa = 3.91NVLSHH211 pKa = 5.41IHH213 pKa = 6.52KK214 pKa = 10.53FLDD217 pKa = 3.5TKK219 pKa = 10.72PYY221 pKa = 10.62LPLHH225 pKa = 5.97FVDD228 pKa = 4.11TQFCKK233 pKa = 10.39TPLVTGTGYY242 pKa = 10.81HH243 pKa = 5.59NRR245 pKa = 11.84YY246 pKa = 9.47SFKK249 pKa = 10.71QKK251 pKa = 10.14AHH253 pKa = 6.25AKK255 pKa = 8.19YY256 pKa = 10.18SHH258 pKa = 6.48PAEE261 pKa = 4.48YY262 pKa = 8.59ATKK265 pKa = 9.37PISKK269 pKa = 10.03GYY271 pKa = 9.88FYY273 pKa = 11.23NATYY277 pKa = 10.76EE278 pKa = 3.93NARR281 pKa = 11.84TIIHH285 pKa = 6.83FIKK288 pKa = 10.54EE289 pKa = 3.72YY290 pKa = 10.52GLPFNVIRR298 pKa = 11.84ADD300 pKa = 3.92DD301 pKa = 3.98KK302 pKa = 11.95SEE304 pKa = 3.99LTDD307 pKa = 3.83SDD309 pKa = 3.74VQKK312 pKa = 11.1YY313 pKa = 10.08INEE316 pKa = 4.19ANSFFNDD323 pKa = 3.31YY324 pKa = 7.74PTLLFTRR331 pKa = 11.84NHH333 pKa = 5.47ISKK336 pKa = 10.27RR337 pKa = 11.84DD338 pKa = 3.55GPLKK342 pKa = 10.16VRR344 pKa = 11.84PVYY347 pKa = 10.62AVDD350 pKa = 4.65DD351 pKa = 3.68IFIIMEE357 pKa = 4.25LMLTFPLTIQARR369 pKa = 11.84KK370 pKa = 8.74PSCCIMYY377 pKa = 10.42GLEE380 pKa = 4.28TIRR383 pKa = 11.84GSNRR387 pKa = 11.84YY388 pKa = 8.8IEE390 pKa = 4.44QIARR394 pKa = 11.84DD395 pKa = 3.74YY396 pKa = 11.09STFFSLDD403 pKa = 2.74WSSYY407 pKa = 6.99DD408 pKa = 3.17QRR410 pKa = 11.84LPRR413 pKa = 11.84VITDD417 pKa = 3.3IYY419 pKa = 9.28YY420 pKa = 9.62TDD422 pKa = 4.27FLRR425 pKa = 11.84SLLVINNGYY434 pKa = 9.28QPTYY438 pKa = 10.24EE439 pKa = 4.22YY440 pKa = 8.37PTYY443 pKa = 10.44PDD445 pKa = 3.72LDD447 pKa = 3.61EE448 pKa = 5.24HH449 pKa = 6.91KK450 pKa = 10.3LYY452 pKa = 11.02HH453 pKa = 7.43RR454 pKa = 11.84MDD456 pKa = 3.79NLLHH460 pKa = 6.85FLHH463 pKa = 6.72LWYY466 pKa = 11.33NNMTFLLPDD475 pKa = 4.16GYY477 pKa = 11.15AYY479 pKa = 10.7RR480 pKa = 11.84RR481 pKa = 11.84TSCGVPSGLYY491 pKa = 7.56NTQYY495 pKa = 11.31LDD497 pKa = 3.35SFGNLFLIIDD507 pKa = 4.03AMLEE511 pKa = 3.96FGFTDD516 pKa = 3.75SEE518 pKa = 4.08IQKK521 pKa = 10.67FILLVLGDD529 pKa = 5.24DD530 pKa = 3.93NTGMTTIPICRR541 pKa = 11.84MFNFITFLEE550 pKa = 4.95KK551 pKa = 10.76YY552 pKa = 10.2ALEE555 pKa = 4.49RR556 pKa = 11.84YY557 pKa = 10.39NMVLSTTKK565 pKa = 10.58SVLTTFRR572 pKa = 11.84SKK574 pKa = 10.49IEE576 pKa = 3.95SLGYY580 pKa = 9.35QCNHH584 pKa = 6.66GSPKK588 pKa = 9.96RR589 pKa = 11.84DD590 pKa = 2.8IDD592 pKa = 3.69KK593 pKa = 11.03LVAQLCYY600 pKa = 9.98PEE602 pKa = 5.79NGLKK606 pKa = 9.45PHH608 pKa = 6.12TMAARR613 pKa = 11.84AIGIAYY619 pKa = 9.85ASAGQDD625 pKa = 3.3DD626 pKa = 4.87MFHH629 pKa = 6.69SFCQDD634 pKa = 2.32VYY636 pKa = 11.4NIFRR640 pKa = 11.84SDD642 pKa = 3.51YY643 pKa = 11.0RR644 pKa = 11.84PDD646 pKa = 3.55DD647 pKa = 3.83RR648 pKa = 11.84MNLHH652 pKa = 7.07FKK654 pKa = 10.58RR655 pKa = 11.84QIFHH659 pKa = 6.44NLEE662 pKa = 4.65DD663 pKa = 4.34GMPDD667 pKa = 3.93LAPPIVPPFPSLYY680 pKa = 10.08EE681 pKa = 3.62IRR683 pKa = 11.84EE684 pKa = 3.76MYY686 pKa = 10.25AYY688 pKa = 10.76YY689 pKa = 10.45KK690 pKa = 10.81GPLDD694 pKa = 5.61FAPKK698 pKa = 8.95WNYY701 pKa = 9.41AHH703 pKa = 6.86FMSDD707 pKa = 3.44PDD709 pKa = 3.77TTPPYY714 pKa = 10.32SKK716 pKa = 9.81TMRR719 pKa = 11.84DD720 pKa = 3.47YY721 pKa = 10.87EE722 pKa = 4.28AEE724 pKa = 4.06NNISSRR730 pKa = 11.84VAPTFEE736 pKa = 4.21TVVPSTINLPP746 pKa = 3.63
MM1 pKa = 7.81PFNAVRR7 pKa = 11.84NYY9 pKa = 10.52LNEE12 pKa = 3.53RR13 pKa = 11.84MTRR16 pKa = 11.84LKK18 pKa = 10.63QEE20 pKa = 3.51WKK22 pKa = 8.49TYY24 pKa = 10.3QSTDD28 pKa = 2.96HH29 pKa = 7.31DD30 pKa = 5.06PIKK33 pKa = 10.71ILDD36 pKa = 4.3TIQDD40 pKa = 3.59PDD42 pKa = 3.32YY43 pKa = 11.23RR44 pKa = 11.84RR45 pKa = 11.84YY46 pKa = 10.67LDD48 pKa = 3.33NARR51 pKa = 11.84FNSDD55 pKa = 3.17NEE57 pKa = 4.48MKK59 pKa = 10.57HH60 pKa = 6.7LMLNKK65 pKa = 9.91EE66 pKa = 3.89YY67 pKa = 9.61STLVEE72 pKa = 5.36AYY74 pKa = 8.28RR75 pKa = 11.84TDD77 pKa = 3.31NAKK80 pKa = 9.94KK81 pKa = 7.82HH82 pKa = 4.88QVYY85 pKa = 9.84EE86 pKa = 3.83LHH88 pKa = 6.59QPIPNDD94 pKa = 3.58AAPVIQSRR102 pKa = 11.84LPAKK106 pKa = 10.1GIKK109 pKa = 9.39LVPLMYY115 pKa = 10.13HH116 pKa = 6.17YY117 pKa = 11.45GHH119 pKa = 6.32VTHH122 pKa = 7.42DD123 pKa = 3.91PVSLTDD129 pKa = 4.02ANRR132 pKa = 11.84SDD134 pKa = 4.71DD135 pKa = 4.44FEE137 pKa = 4.61TQVVPDD143 pKa = 3.73SDD145 pKa = 3.55APTRR149 pKa = 11.84FGYY152 pKa = 10.0PIDD155 pKa = 3.56VRR157 pKa = 11.84IYY159 pKa = 10.33NLICYY164 pKa = 9.56RR165 pKa = 11.84YY166 pKa = 8.73SKK168 pKa = 10.59YY169 pKa = 11.01LEE171 pKa = 4.51VINAYY176 pKa = 9.52CRR178 pKa = 11.84PIGTVNATFEE188 pKa = 4.58DD189 pKa = 4.7FNKK192 pKa = 10.12EE193 pKa = 4.12QIPSAPIDD201 pKa = 3.67PKK203 pKa = 10.73RR204 pKa = 11.84KK205 pKa = 9.58EE206 pKa = 3.91NVLSHH211 pKa = 5.41IHH213 pKa = 6.52KK214 pKa = 10.53FLDD217 pKa = 3.5TKK219 pKa = 10.72PYY221 pKa = 10.62LPLHH225 pKa = 5.97FVDD228 pKa = 4.11TQFCKK233 pKa = 10.39TPLVTGTGYY242 pKa = 10.81HH243 pKa = 5.59NRR245 pKa = 11.84YY246 pKa = 9.47SFKK249 pKa = 10.71QKK251 pKa = 10.14AHH253 pKa = 6.25AKK255 pKa = 8.19YY256 pKa = 10.18SHH258 pKa = 6.48PAEE261 pKa = 4.48YY262 pKa = 8.59ATKK265 pKa = 9.37PISKK269 pKa = 10.03GYY271 pKa = 9.88FYY273 pKa = 11.23NATYY277 pKa = 10.76EE278 pKa = 3.93NARR281 pKa = 11.84TIIHH285 pKa = 6.83FIKK288 pKa = 10.54EE289 pKa = 3.72YY290 pKa = 10.52GLPFNVIRR298 pKa = 11.84ADD300 pKa = 3.92DD301 pKa = 3.98KK302 pKa = 11.95SEE304 pKa = 3.99LTDD307 pKa = 3.83SDD309 pKa = 3.74VQKK312 pKa = 11.1YY313 pKa = 10.08INEE316 pKa = 4.19ANSFFNDD323 pKa = 3.31YY324 pKa = 7.74PTLLFTRR331 pKa = 11.84NHH333 pKa = 5.47ISKK336 pKa = 10.27RR337 pKa = 11.84DD338 pKa = 3.55GPLKK342 pKa = 10.16VRR344 pKa = 11.84PVYY347 pKa = 10.62AVDD350 pKa = 4.65DD351 pKa = 3.68IFIIMEE357 pKa = 4.25LMLTFPLTIQARR369 pKa = 11.84KK370 pKa = 8.74PSCCIMYY377 pKa = 10.42GLEE380 pKa = 4.28TIRR383 pKa = 11.84GSNRR387 pKa = 11.84YY388 pKa = 8.8IEE390 pKa = 4.44QIARR394 pKa = 11.84DD395 pKa = 3.74YY396 pKa = 11.09STFFSLDD403 pKa = 2.74WSSYY407 pKa = 6.99DD408 pKa = 3.17QRR410 pKa = 11.84LPRR413 pKa = 11.84VITDD417 pKa = 3.3IYY419 pKa = 9.28YY420 pKa = 9.62TDD422 pKa = 4.27FLRR425 pKa = 11.84SLLVINNGYY434 pKa = 9.28QPTYY438 pKa = 10.24EE439 pKa = 4.22YY440 pKa = 8.37PTYY443 pKa = 10.44PDD445 pKa = 3.72LDD447 pKa = 3.61EE448 pKa = 5.24HH449 pKa = 6.91KK450 pKa = 10.3LYY452 pKa = 11.02HH453 pKa = 7.43RR454 pKa = 11.84MDD456 pKa = 3.79NLLHH460 pKa = 6.85FLHH463 pKa = 6.72LWYY466 pKa = 11.33NNMTFLLPDD475 pKa = 4.16GYY477 pKa = 11.15AYY479 pKa = 10.7RR480 pKa = 11.84RR481 pKa = 11.84TSCGVPSGLYY491 pKa = 7.56NTQYY495 pKa = 11.31LDD497 pKa = 3.35SFGNLFLIIDD507 pKa = 4.03AMLEE511 pKa = 3.96FGFTDD516 pKa = 3.75SEE518 pKa = 4.08IQKK521 pKa = 10.67FILLVLGDD529 pKa = 5.24DD530 pKa = 3.93NTGMTTIPICRR541 pKa = 11.84MFNFITFLEE550 pKa = 4.95KK551 pKa = 10.76YY552 pKa = 10.2ALEE555 pKa = 4.49RR556 pKa = 11.84YY557 pKa = 10.39NMVLSTTKK565 pKa = 10.58SVLTTFRR572 pKa = 11.84SKK574 pKa = 10.49IEE576 pKa = 3.95SLGYY580 pKa = 9.35QCNHH584 pKa = 6.66GSPKK588 pKa = 9.96RR589 pKa = 11.84DD590 pKa = 2.8IDD592 pKa = 3.69KK593 pKa = 11.03LVAQLCYY600 pKa = 9.98PEE602 pKa = 5.79NGLKK606 pKa = 9.45PHH608 pKa = 6.12TMAARR613 pKa = 11.84AIGIAYY619 pKa = 9.85ASAGQDD625 pKa = 3.3DD626 pKa = 4.87MFHH629 pKa = 6.69SFCQDD634 pKa = 2.32VYY636 pKa = 11.4NIFRR640 pKa = 11.84SDD642 pKa = 3.51YY643 pKa = 11.0RR644 pKa = 11.84PDD646 pKa = 3.55DD647 pKa = 3.83RR648 pKa = 11.84MNLHH652 pKa = 7.07FKK654 pKa = 10.58RR655 pKa = 11.84QIFHH659 pKa = 6.44NLEE662 pKa = 4.65DD663 pKa = 4.34GMPDD667 pKa = 3.93LAPPIVPPFPSLYY680 pKa = 10.08EE681 pKa = 3.62IRR683 pKa = 11.84EE684 pKa = 3.76MYY686 pKa = 10.25AYY688 pKa = 10.76YY689 pKa = 10.45KK690 pKa = 10.81GPLDD694 pKa = 5.61FAPKK698 pKa = 8.95WNYY701 pKa = 9.41AHH703 pKa = 6.86FMSDD707 pKa = 3.44PDD709 pKa = 3.77TTPPYY714 pKa = 10.32SKK716 pKa = 9.81TMRR719 pKa = 11.84DD720 pKa = 3.47YY721 pKa = 10.87EE722 pKa = 4.28AEE724 pKa = 4.06NNISSRR730 pKa = 11.84VAPTFEE736 pKa = 4.21TVVPSTINLPP746 pKa = 3.63
Molecular weight: 87.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1420 |
674 |
746 |
710.0 |
81.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.042 ± 1.352 | 1.127 ± 0.159 |
6.479 ± 0.766 | 4.225 ± 0.148 |
5.634 ± 0.103 | 3.31 ± 0.131 |
3.521 ± 0.127 | 5.915 ± 0.487 |
4.155 ± 0.8 | 8.944 ± 0.372 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.324 ± 0.366 | 6.127 ± 0.27 |
7.254 ± 0.211 | 2.676 ± 0.203 |
5.282 ± 0.26 | 6.761 ± 0.843 |
7.676 ± 0.726 | 5.0 ± 0.53 |
0.634 ± 0.073 | 5.915 ± 1.286 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
