
Methylotenera mobilis (strain JLW8 / ATCC BAA-1282 / DSM 17540)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Methylophilaceae; Methylotenera; Methylotenera mobilis
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
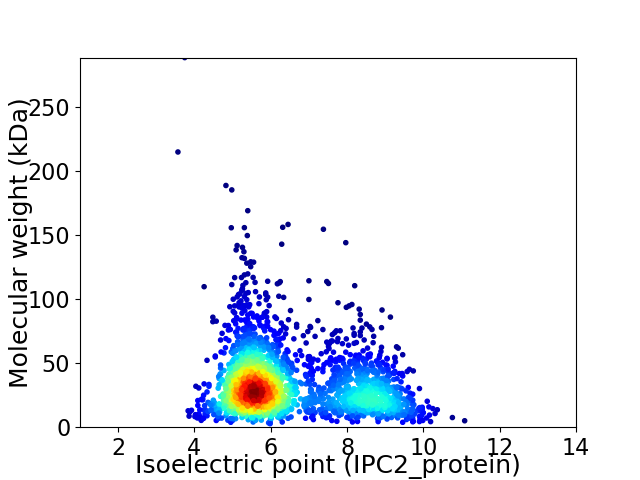
Virtual 2D-PAGE plot for 2334 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
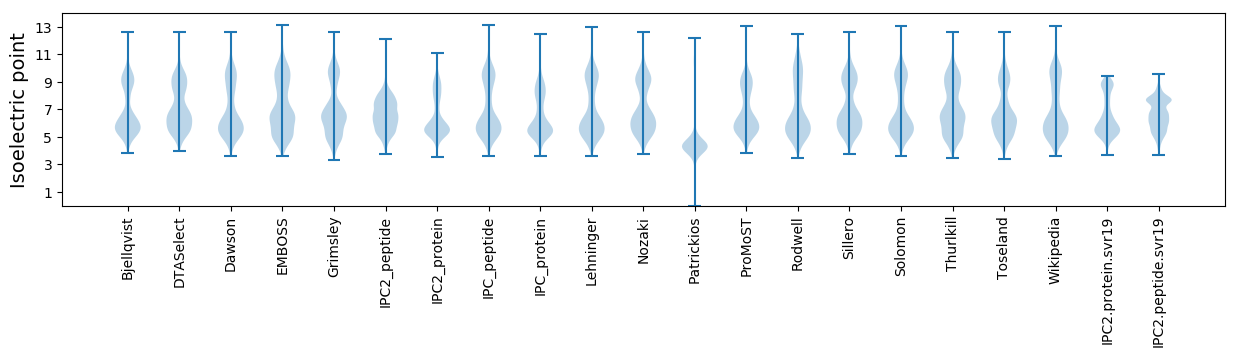
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C6WV09|C6WV09_METML Uncharacterized protein OS=Methylotenera mobilis (strain JLW8 / ATCC BAA-1282 / DSM 17540) OX=583345 GN=Mmol_0848 PE=4 SV=1
MM1 pKa = 7.48ALIGKK6 pKa = 8.81IIAMTGTASLISNNGNQRR24 pKa = 11.84DD25 pKa = 4.01LRR27 pKa = 11.84LGDD30 pKa = 4.37NIQTADD36 pKa = 4.0TIKK39 pKa = 9.69TGAGVEE45 pKa = 4.08VDD47 pKa = 3.67LQLANGQVVHH57 pKa = 7.02IGAEE61 pKa = 3.99QLVAFADD68 pKa = 3.88EE69 pKa = 4.34LVEE72 pKa = 4.54DD73 pKa = 5.74FIPATLDD80 pKa = 3.1TSVNVATIDD89 pKa = 3.78TVVKK93 pKa = 10.57AIEE96 pKa = 4.08EE97 pKa = 4.33GKK99 pKa = 10.59DD100 pKa = 2.94IGDD103 pKa = 3.76VLEE106 pKa = 4.44EE107 pKa = 4.03TAAGPGGSSNSYY119 pKa = 9.86GFSFVDD125 pKa = 3.93LLRR128 pKa = 11.84INDD131 pKa = 3.93DD132 pKa = 3.65LNNFRR137 pKa = 11.84FGYY140 pKa = 9.6EE141 pKa = 4.14FGSATPNEE149 pKa = 4.2QALSGNSFADD159 pKa = 3.34TLNQINVVTGTGGAPDD175 pKa = 4.01TNSPSVVVNIVDD187 pKa = 3.67ANLNVADD194 pKa = 4.16TVSNVTFTFSEE205 pKa = 4.6APVGFTIGDD214 pKa = 3.37ISSTGGIISNLVATADD230 pKa = 3.85PLVFTATFTANAGFEE245 pKa = 4.54GTGSVSVNAGSYY257 pKa = 9.55TNASGNVGTSGADD270 pKa = 3.08TVGIDD275 pKa = 3.36TAAPVPTITLTPNVTADD292 pKa = 4.06DD293 pKa = 4.6IINAGEE299 pKa = 3.88AGGNVNITGTVGGDD313 pKa = 2.91AKK315 pKa = 11.58VNDD318 pKa = 4.13TVTLVINGNTYY329 pKa = 9.85TGLVQPGLTFSIPVAGSDD347 pKa = 4.57LVADD351 pKa = 4.45PDD353 pKa = 3.89RR354 pKa = 11.84VIDD357 pKa = 4.08ASVSTTDD364 pKa = 3.63AAGNTATASDD374 pKa = 4.0TEE376 pKa = 4.74GYY378 pKa = 10.15SVDD381 pKa = 3.54STVPTIANQTFSYY394 pKa = 10.76AEE396 pKa = 3.76NRR398 pKa = 11.84AAGATVATVVASDD411 pKa = 3.54NVGVTGYY418 pKa = 10.88SFTATGTNTSADD430 pKa = 3.41GYY432 pKa = 8.73YY433 pKa = 10.11QISNTGVITITAAGAASGVNDD454 pKa = 4.43FEE456 pKa = 5.1QGTNTGNYY464 pKa = 9.24GITVRR469 pKa = 11.84DD470 pKa = 3.3AAGNNSNATITLNEE484 pKa = 4.07TDD486 pKa = 5.47LDD488 pKa = 3.89DD489 pKa = 4.5TNPVIANQTFSYY501 pKa = 10.76AEE503 pKa = 3.76NRR505 pKa = 11.84AAGATVATVVASDD518 pKa = 3.54NVGVTGYY525 pKa = 10.88SFTATGTNTSADD537 pKa = 3.41GYY539 pKa = 8.73YY540 pKa = 10.11QISNTGVITITAAGAASGVNDD561 pKa = 4.43FEE563 pKa = 5.1QGTNTGNYY571 pKa = 9.24GITVRR576 pKa = 11.84DD577 pKa = 3.3AAGNNSNATITLNEE591 pKa = 4.25TNVNEE596 pKa = 3.96PVAAVSDD603 pKa = 4.45AYY605 pKa = 10.0TMNEE609 pKa = 4.33DD610 pKa = 3.63GPAITLTPLANDD622 pKa = 3.81SDD624 pKa = 4.89PDD626 pKa = 3.64GTTPTIQSINGTALTPGVAQAIAVTGGMVNVSAAGVITFTPTLNFNGTVTFPYY679 pKa = 10.36VITDD683 pKa = 4.07GATTATANQVITVNAVDD700 pKa = 4.61DD701 pKa = 4.48PSVLIADD708 pKa = 4.11TNTVNEE714 pKa = 4.77DD715 pKa = 3.15NPATGNVLSNDD726 pKa = 3.29SDD728 pKa = 3.66VDD730 pKa = 3.66NVLTVSSFKK739 pKa = 10.88VAGISGTFTAGQTATITGVGTLTIAANGNYY769 pKa = 9.01TFTPVANYY777 pKa = 9.91NGSVPVATYY786 pKa = 8.82TVNTGSTSTLSITVTPVNDD805 pKa = 3.72APVGVADD812 pKa = 4.07VFNGVNSVVEE822 pKa = 4.15GTTVVRR828 pKa = 11.84GNILANDD835 pKa = 3.95TDD837 pKa = 3.85VDD839 pKa = 4.26STTLTVAQFATNSGATATAVNGTNSITTALGGTVVMNADD878 pKa = 3.3GTFTYY883 pKa = 7.9TAPVRR888 pKa = 11.84NHH890 pKa = 6.55ADD892 pKa = 3.73AISDD896 pKa = 3.27VDD898 pKa = 3.97SFVYY902 pKa = 10.3RR903 pKa = 11.84ATDD906 pKa = 3.57GSLNSAWTTVTLNIADD922 pKa = 4.25SVPVANPDD930 pKa = 3.4TDD932 pKa = 3.84SVGVGFRR939 pKa = 11.84NPTTNNTASVTGNVITGAGDD959 pKa = 3.2TGGVDD964 pKa = 3.43TLGADD969 pKa = 3.7AARR972 pKa = 11.84VSSIVFNGTTYY983 pKa = 11.29NLSAGNTSIVTSNGTLLINEE1003 pKa = 4.54TGSYY1007 pKa = 9.6TYY1009 pKa = 10.42TSSYY1013 pKa = 8.19QNKK1016 pKa = 8.23VVPVSPSNTASTVANWTSAGISSFGFDD1043 pKa = 3.08GTSPLTNSNNTLTLSALTTAASNIVRR1069 pKa = 11.84LSDD1072 pKa = 3.38NTNSNNDD1079 pKa = 3.94GIGVEE1084 pKa = 4.48TTLSGSASNQSNISRR1099 pKa = 11.84IEE1101 pKa = 4.08NNEE1104 pKa = 3.9HH1105 pKa = 6.54LVLNLGMLSRR1115 pKa = 11.84NTSVTLTDD1123 pKa = 3.82LTASEE1128 pKa = 4.31TAQWRR1133 pKa = 11.84AYY1135 pKa = 9.19DD1136 pKa = 3.6ANGAIVASGTITGNSTNIATGTVSSTVAFQYY1167 pKa = 10.64IVFSGSSSANFRR1179 pKa = 11.84VNGLTATPDD1188 pKa = 3.54LTAVTPDD1195 pKa = 2.85QFTYY1199 pKa = 10.78TLTDD1203 pKa = 3.44ADD1205 pKa = 4.34GSTASTTLTINTNGNPSAVADD1226 pKa = 3.66SATVYY1231 pKa = 10.71EE1232 pKa = 4.81SGLSTGTQAGVLATTASGNLLVNDD1256 pKa = 4.8AGVSTTTTISSINGVTPTSGTITVTSATGTLVVNANTGAYY1296 pKa = 8.22TYY1298 pKa = 9.38TLNAATTEE1306 pKa = 4.19GVNDD1310 pKa = 3.96KK1311 pKa = 9.18PTFNYY1316 pKa = 10.29VLTDD1320 pKa = 3.51SVTGQSTNANLTVNIVDD1337 pKa = 4.23DD1338 pKa = 4.82APVGGNITQTLQAASAALTYY1358 pKa = 10.91NVVIVLDD1365 pKa = 4.22RR1366 pKa = 11.84SGSMAQDD1373 pKa = 3.95ANGLWSNQSGYY1384 pKa = 10.63DD1385 pKa = 3.17ASTNRR1390 pKa = 11.84MEE1392 pKa = 4.27IAKK1395 pKa = 9.79EE1396 pKa = 3.91AIAQLIARR1404 pKa = 11.84YY1405 pKa = 9.86DD1406 pKa = 3.39GLGNVNVKK1414 pKa = 9.95FVTFSSDD1421 pKa = 2.98AVEE1424 pKa = 4.33SEE1426 pKa = 4.06WYY1428 pKa = 9.39IDD1430 pKa = 3.59NVTGAVRR1437 pKa = 11.84YY1438 pKa = 9.54VDD1440 pKa = 3.69NVQAGGGTQYY1450 pKa = 10.0STALNEE1456 pKa = 4.26TMSGFTQPVADD1467 pKa = 3.44KK1468 pKa = 10.02TLFYY1472 pKa = 10.46FITDD1476 pKa = 4.05GQPNSGYY1483 pKa = 10.39EE1484 pKa = 3.87VDD1486 pKa = 5.13ATLQTQWQNFVAANGNISFGIGIGTASLSSLTPIAYY1522 pKa = 9.22PNVDD1526 pKa = 3.02ADD1528 pKa = 4.18GNGTEE1533 pKa = 5.01DD1534 pKa = 3.4YY1535 pKa = 10.6AIRR1538 pKa = 11.84VDD1540 pKa = 4.2NPADD1544 pKa = 3.65LADD1547 pKa = 4.13TLLSTVDD1554 pKa = 3.29GGVVVGNMSVLSGSGSSGFLLGADD1578 pKa = 3.9GGRR1581 pKa = 11.84LNSVVVDD1588 pKa = 4.16GVTYY1592 pKa = 9.21TYY1594 pKa = 10.66SASGPSSMTISTNKK1608 pKa = 10.24GGEE1611 pKa = 4.04LTVNFLTGEE1620 pKa = 4.08YY1621 pKa = 9.93SYY1623 pKa = 11.6HH1624 pKa = 6.58LLLNRR1629 pKa = 11.84TVQNEE1634 pKa = 4.1QEE1636 pKa = 4.5SFLVTAVDD1644 pKa = 3.76GDD1646 pKa = 4.23GDD1648 pKa = 4.04TKK1650 pKa = 10.85TINLNINLDD1659 pKa = 3.81YY1660 pKa = 11.02VANLDD1665 pKa = 4.02PNADD1669 pKa = 3.98TILTNVVTGTPISVSAAALLHH1690 pKa = 6.05NDD1692 pKa = 3.52SLRR1695 pKa = 11.84GTGSITSTQNAVNGSVSGTSTVQFTANAVAAKK1727 pKa = 8.33TIQVVTEE1734 pKa = 3.99ASFDD1738 pKa = 3.94STSQTQNSTRR1748 pKa = 11.84EE1749 pKa = 4.0NAVDD1753 pKa = 3.35LTDD1756 pKa = 3.52RR1757 pKa = 11.84SKK1759 pKa = 11.2FGTVVSGTPTWAVDD1773 pKa = 3.49VAGGSSIVFSGLLDD1787 pKa = 3.55NSGSGTRR1794 pKa = 11.84DD1795 pKa = 2.64VDD1797 pKa = 3.82YY1798 pKa = 11.52VKK1800 pKa = 11.12VKK1802 pKa = 9.21MYY1804 pKa = 10.46AGEE1807 pKa = 4.09RR1808 pKa = 11.84VFIDD1812 pKa = 4.19VDD1814 pKa = 3.58NQTQGVNAFVEE1825 pKa = 4.46YY1826 pKa = 10.46FDD1828 pKa = 5.65ANGVLQTFTVGTTGTGTSLAPNGYY1852 pKa = 8.02FTAPEE1857 pKa = 3.9SGEE1860 pKa = 3.88YY1861 pKa = 9.55FIRR1864 pKa = 11.84MQTVGTTDD1872 pKa = 2.79TSYY1875 pKa = 11.96NLVLTLTDD1883 pKa = 2.9IVGPMTEE1890 pKa = 4.29NGSFEE1895 pKa = 4.3YY1896 pKa = 10.26TVTEE1900 pKa = 3.95NGVASSASATVYY1912 pKa = 10.5NVVGNTITGGDD1923 pKa = 3.65GDD1925 pKa = 4.83EE1926 pKa = 4.6ILLGGNTNDD1935 pKa = 3.64TLIGGGGNDD1944 pKa = 3.64VLIGGAGNDD1953 pKa = 3.71TLLGGTGADD1962 pKa = 3.34RR1963 pKa = 11.84LEE1965 pKa = 4.64GGSGNDD1971 pKa = 3.32ILNGGTGNDD1980 pKa = 3.21ILIGGTGNDD1989 pKa = 3.89TLTGGLGADD1998 pKa = 3.47VFKK2001 pKa = 10.84WSLADD2006 pKa = 3.89AGATGTPATDD2016 pKa = 3.57VITDD2020 pKa = 4.32FDD2022 pKa = 3.84TTANSDD2028 pKa = 3.72KK2029 pKa = 11.21LDD2031 pKa = 4.69LRR2033 pKa = 11.84DD2034 pKa = 4.03LLQGEE2039 pKa = 4.48IGSGVGANLEE2049 pKa = 4.27NYY2051 pKa = 10.32LHH2053 pKa = 6.44FEE2055 pKa = 4.12KK2056 pKa = 10.91VGTDD2060 pKa = 3.39TVVHH2064 pKa = 6.59ISSNGSFNNGYY2075 pKa = 9.73NPAAEE2080 pKa = 4.3VQTITLQNVDD2090 pKa = 4.11LVGSYY2095 pKa = 11.52ANDD2098 pKa = 3.22QQVIQNLINNQKK2110 pKa = 10.63LITDD2114 pKa = 4.23
MM1 pKa = 7.48ALIGKK6 pKa = 8.81IIAMTGTASLISNNGNQRR24 pKa = 11.84DD25 pKa = 4.01LRR27 pKa = 11.84LGDD30 pKa = 4.37NIQTADD36 pKa = 4.0TIKK39 pKa = 9.69TGAGVEE45 pKa = 4.08VDD47 pKa = 3.67LQLANGQVVHH57 pKa = 7.02IGAEE61 pKa = 3.99QLVAFADD68 pKa = 3.88EE69 pKa = 4.34LVEE72 pKa = 4.54DD73 pKa = 5.74FIPATLDD80 pKa = 3.1TSVNVATIDD89 pKa = 3.78TVVKK93 pKa = 10.57AIEE96 pKa = 4.08EE97 pKa = 4.33GKK99 pKa = 10.59DD100 pKa = 2.94IGDD103 pKa = 3.76VLEE106 pKa = 4.44EE107 pKa = 4.03TAAGPGGSSNSYY119 pKa = 9.86GFSFVDD125 pKa = 3.93LLRR128 pKa = 11.84INDD131 pKa = 3.93DD132 pKa = 3.65LNNFRR137 pKa = 11.84FGYY140 pKa = 9.6EE141 pKa = 4.14FGSATPNEE149 pKa = 4.2QALSGNSFADD159 pKa = 3.34TLNQINVVTGTGGAPDD175 pKa = 4.01TNSPSVVVNIVDD187 pKa = 3.67ANLNVADD194 pKa = 4.16TVSNVTFTFSEE205 pKa = 4.6APVGFTIGDD214 pKa = 3.37ISSTGGIISNLVATADD230 pKa = 3.85PLVFTATFTANAGFEE245 pKa = 4.54GTGSVSVNAGSYY257 pKa = 9.55TNASGNVGTSGADD270 pKa = 3.08TVGIDD275 pKa = 3.36TAAPVPTITLTPNVTADD292 pKa = 4.06DD293 pKa = 4.6IINAGEE299 pKa = 3.88AGGNVNITGTVGGDD313 pKa = 2.91AKK315 pKa = 11.58VNDD318 pKa = 4.13TVTLVINGNTYY329 pKa = 9.85TGLVQPGLTFSIPVAGSDD347 pKa = 4.57LVADD351 pKa = 4.45PDD353 pKa = 3.89RR354 pKa = 11.84VIDD357 pKa = 4.08ASVSTTDD364 pKa = 3.63AAGNTATASDD374 pKa = 4.0TEE376 pKa = 4.74GYY378 pKa = 10.15SVDD381 pKa = 3.54STVPTIANQTFSYY394 pKa = 10.76AEE396 pKa = 3.76NRR398 pKa = 11.84AAGATVATVVASDD411 pKa = 3.54NVGVTGYY418 pKa = 10.88SFTATGTNTSADD430 pKa = 3.41GYY432 pKa = 8.73YY433 pKa = 10.11QISNTGVITITAAGAASGVNDD454 pKa = 4.43FEE456 pKa = 5.1QGTNTGNYY464 pKa = 9.24GITVRR469 pKa = 11.84DD470 pKa = 3.3AAGNNSNATITLNEE484 pKa = 4.07TDD486 pKa = 5.47LDD488 pKa = 3.89DD489 pKa = 4.5TNPVIANQTFSYY501 pKa = 10.76AEE503 pKa = 3.76NRR505 pKa = 11.84AAGATVATVVASDD518 pKa = 3.54NVGVTGYY525 pKa = 10.88SFTATGTNTSADD537 pKa = 3.41GYY539 pKa = 8.73YY540 pKa = 10.11QISNTGVITITAAGAASGVNDD561 pKa = 4.43FEE563 pKa = 5.1QGTNTGNYY571 pKa = 9.24GITVRR576 pKa = 11.84DD577 pKa = 3.3AAGNNSNATITLNEE591 pKa = 4.25TNVNEE596 pKa = 3.96PVAAVSDD603 pKa = 4.45AYY605 pKa = 10.0TMNEE609 pKa = 4.33DD610 pKa = 3.63GPAITLTPLANDD622 pKa = 3.81SDD624 pKa = 4.89PDD626 pKa = 3.64GTTPTIQSINGTALTPGVAQAIAVTGGMVNVSAAGVITFTPTLNFNGTVTFPYY679 pKa = 10.36VITDD683 pKa = 4.07GATTATANQVITVNAVDD700 pKa = 4.61DD701 pKa = 4.48PSVLIADD708 pKa = 4.11TNTVNEE714 pKa = 4.77DD715 pKa = 3.15NPATGNVLSNDD726 pKa = 3.29SDD728 pKa = 3.66VDD730 pKa = 3.66NVLTVSSFKK739 pKa = 10.88VAGISGTFTAGQTATITGVGTLTIAANGNYY769 pKa = 9.01TFTPVANYY777 pKa = 9.91NGSVPVATYY786 pKa = 8.82TVNTGSTSTLSITVTPVNDD805 pKa = 3.72APVGVADD812 pKa = 4.07VFNGVNSVVEE822 pKa = 4.15GTTVVRR828 pKa = 11.84GNILANDD835 pKa = 3.95TDD837 pKa = 3.85VDD839 pKa = 4.26STTLTVAQFATNSGATATAVNGTNSITTALGGTVVMNADD878 pKa = 3.3GTFTYY883 pKa = 7.9TAPVRR888 pKa = 11.84NHH890 pKa = 6.55ADD892 pKa = 3.73AISDD896 pKa = 3.27VDD898 pKa = 3.97SFVYY902 pKa = 10.3RR903 pKa = 11.84ATDD906 pKa = 3.57GSLNSAWTTVTLNIADD922 pKa = 4.25SVPVANPDD930 pKa = 3.4TDD932 pKa = 3.84SVGVGFRR939 pKa = 11.84NPTTNNTASVTGNVITGAGDD959 pKa = 3.2TGGVDD964 pKa = 3.43TLGADD969 pKa = 3.7AARR972 pKa = 11.84VSSIVFNGTTYY983 pKa = 11.29NLSAGNTSIVTSNGTLLINEE1003 pKa = 4.54TGSYY1007 pKa = 9.6TYY1009 pKa = 10.42TSSYY1013 pKa = 8.19QNKK1016 pKa = 8.23VVPVSPSNTASTVANWTSAGISSFGFDD1043 pKa = 3.08GTSPLTNSNNTLTLSALTTAASNIVRR1069 pKa = 11.84LSDD1072 pKa = 3.38NTNSNNDD1079 pKa = 3.94GIGVEE1084 pKa = 4.48TTLSGSASNQSNISRR1099 pKa = 11.84IEE1101 pKa = 4.08NNEE1104 pKa = 3.9HH1105 pKa = 6.54LVLNLGMLSRR1115 pKa = 11.84NTSVTLTDD1123 pKa = 3.82LTASEE1128 pKa = 4.31TAQWRR1133 pKa = 11.84AYY1135 pKa = 9.19DD1136 pKa = 3.6ANGAIVASGTITGNSTNIATGTVSSTVAFQYY1167 pKa = 10.64IVFSGSSSANFRR1179 pKa = 11.84VNGLTATPDD1188 pKa = 3.54LTAVTPDD1195 pKa = 2.85QFTYY1199 pKa = 10.78TLTDD1203 pKa = 3.44ADD1205 pKa = 4.34GSTASTTLTINTNGNPSAVADD1226 pKa = 3.66SATVYY1231 pKa = 10.71EE1232 pKa = 4.81SGLSTGTQAGVLATTASGNLLVNDD1256 pKa = 4.8AGVSTTTTISSINGVTPTSGTITVTSATGTLVVNANTGAYY1296 pKa = 8.22TYY1298 pKa = 9.38TLNAATTEE1306 pKa = 4.19GVNDD1310 pKa = 3.96KK1311 pKa = 9.18PTFNYY1316 pKa = 10.29VLTDD1320 pKa = 3.51SVTGQSTNANLTVNIVDD1337 pKa = 4.23DD1338 pKa = 4.82APVGGNITQTLQAASAALTYY1358 pKa = 10.91NVVIVLDD1365 pKa = 4.22RR1366 pKa = 11.84SGSMAQDD1373 pKa = 3.95ANGLWSNQSGYY1384 pKa = 10.63DD1385 pKa = 3.17ASTNRR1390 pKa = 11.84MEE1392 pKa = 4.27IAKK1395 pKa = 9.79EE1396 pKa = 3.91AIAQLIARR1404 pKa = 11.84YY1405 pKa = 9.86DD1406 pKa = 3.39GLGNVNVKK1414 pKa = 9.95FVTFSSDD1421 pKa = 2.98AVEE1424 pKa = 4.33SEE1426 pKa = 4.06WYY1428 pKa = 9.39IDD1430 pKa = 3.59NVTGAVRR1437 pKa = 11.84YY1438 pKa = 9.54VDD1440 pKa = 3.69NVQAGGGTQYY1450 pKa = 10.0STALNEE1456 pKa = 4.26TMSGFTQPVADD1467 pKa = 3.44KK1468 pKa = 10.02TLFYY1472 pKa = 10.46FITDD1476 pKa = 4.05GQPNSGYY1483 pKa = 10.39EE1484 pKa = 3.87VDD1486 pKa = 5.13ATLQTQWQNFVAANGNISFGIGIGTASLSSLTPIAYY1522 pKa = 9.22PNVDD1526 pKa = 3.02ADD1528 pKa = 4.18GNGTEE1533 pKa = 5.01DD1534 pKa = 3.4YY1535 pKa = 10.6AIRR1538 pKa = 11.84VDD1540 pKa = 4.2NPADD1544 pKa = 3.65LADD1547 pKa = 4.13TLLSTVDD1554 pKa = 3.29GGVVVGNMSVLSGSGSSGFLLGADD1578 pKa = 3.9GGRR1581 pKa = 11.84LNSVVVDD1588 pKa = 4.16GVTYY1592 pKa = 9.21TYY1594 pKa = 10.66SASGPSSMTISTNKK1608 pKa = 10.24GGEE1611 pKa = 4.04LTVNFLTGEE1620 pKa = 4.08YY1621 pKa = 9.93SYY1623 pKa = 11.6HH1624 pKa = 6.58LLLNRR1629 pKa = 11.84TVQNEE1634 pKa = 4.1QEE1636 pKa = 4.5SFLVTAVDD1644 pKa = 3.76GDD1646 pKa = 4.23GDD1648 pKa = 4.04TKK1650 pKa = 10.85TINLNINLDD1659 pKa = 3.81YY1660 pKa = 11.02VANLDD1665 pKa = 4.02PNADD1669 pKa = 3.98TILTNVVTGTPISVSAAALLHH1690 pKa = 6.05NDD1692 pKa = 3.52SLRR1695 pKa = 11.84GTGSITSTQNAVNGSVSGTSTVQFTANAVAAKK1727 pKa = 8.33TIQVVTEE1734 pKa = 3.99ASFDD1738 pKa = 3.94STSQTQNSTRR1748 pKa = 11.84EE1749 pKa = 4.0NAVDD1753 pKa = 3.35LTDD1756 pKa = 3.52RR1757 pKa = 11.84SKK1759 pKa = 11.2FGTVVSGTPTWAVDD1773 pKa = 3.49VAGGSSIVFSGLLDD1787 pKa = 3.55NSGSGTRR1794 pKa = 11.84DD1795 pKa = 2.64VDD1797 pKa = 3.82YY1798 pKa = 11.52VKK1800 pKa = 11.12VKK1802 pKa = 9.21MYY1804 pKa = 10.46AGEE1807 pKa = 4.09RR1808 pKa = 11.84VFIDD1812 pKa = 4.19VDD1814 pKa = 3.58NQTQGVNAFVEE1825 pKa = 4.46YY1826 pKa = 10.46FDD1828 pKa = 5.65ANGVLQTFTVGTTGTGTSLAPNGYY1852 pKa = 8.02FTAPEE1857 pKa = 3.9SGEE1860 pKa = 3.88YY1861 pKa = 9.55FIRR1864 pKa = 11.84MQTVGTTDD1872 pKa = 2.79TSYY1875 pKa = 11.96NLVLTLTDD1883 pKa = 2.9IVGPMTEE1890 pKa = 4.29NGSFEE1895 pKa = 4.3YY1896 pKa = 10.26TVTEE1900 pKa = 3.95NGVASSASATVYY1912 pKa = 10.5NVVGNTITGGDD1923 pKa = 3.65GDD1925 pKa = 4.83EE1926 pKa = 4.6ILLGGNTNDD1935 pKa = 3.64TLIGGGGNDD1944 pKa = 3.64VLIGGAGNDD1953 pKa = 3.71TLLGGTGADD1962 pKa = 3.34RR1963 pKa = 11.84LEE1965 pKa = 4.64GGSGNDD1971 pKa = 3.32ILNGGTGNDD1980 pKa = 3.21ILIGGTGNDD1989 pKa = 3.89TLTGGLGADD1998 pKa = 3.47VFKK2001 pKa = 10.84WSLADD2006 pKa = 3.89AGATGTPATDD2016 pKa = 3.57VITDD2020 pKa = 4.32FDD2022 pKa = 3.84TTANSDD2028 pKa = 3.72KK2029 pKa = 11.21LDD2031 pKa = 4.69LRR2033 pKa = 11.84DD2034 pKa = 4.03LLQGEE2039 pKa = 4.48IGSGVGANLEE2049 pKa = 4.27NYY2051 pKa = 10.32LHH2053 pKa = 6.44FEE2055 pKa = 4.12KK2056 pKa = 10.91VGTDD2060 pKa = 3.39TVVHH2064 pKa = 6.59ISSNGSFNNGYY2075 pKa = 9.73NPAAEE2080 pKa = 4.3VQTITLQNVDD2090 pKa = 4.11LVGSYY2095 pKa = 11.52ANDD2098 pKa = 3.22QQVIQNLINNQKK2110 pKa = 10.63LITDD2114 pKa = 4.23
Molecular weight: 214.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6WTX1|C6WTX1_METML Uncharacterized protein OS=Methylotenera mobilis (strain JLW8 / ATCC BAA-1282 / DSM 17540) OX=583345 GN=Mmol_0460 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.25QPSKK9 pKa = 7.79TRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.79GFLVRR21 pKa = 11.84MKK23 pKa = 9.4TKK25 pKa = 10.36GGRR28 pKa = 11.84AVIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.15GRR39 pKa = 11.84SRR41 pKa = 11.84LGLL44 pKa = 3.62
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.25QPSKK9 pKa = 7.79TRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.79GFLVRR21 pKa = 11.84MKK23 pKa = 9.4TKK25 pKa = 10.36GGRR28 pKa = 11.84AVIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.15GRR39 pKa = 11.84SRR41 pKa = 11.84LGLL44 pKa = 3.62
Molecular weight: 5.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
757536 |
23 |
2787 |
324.6 |
35.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.12 ± 0.061 | 0.868 ± 0.019 |
5.273 ± 0.041 | 5.659 ± 0.046 |
3.809 ± 0.035 | 6.82 ± 0.058 |
2.329 ± 0.029 | 6.3 ± 0.044 |
5.375 ± 0.044 | 10.506 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.618 ± 0.025 | 4.353 ± 0.046 |
4.033 ± 0.032 | 4.371 ± 0.038 |
4.635 ± 0.037 | 6.22 ± 0.039 |
5.517 ± 0.046 | 7.202 ± 0.041 |
1.127 ± 0.021 | 2.864 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
