
Streptococcus satellite phage Javan30
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
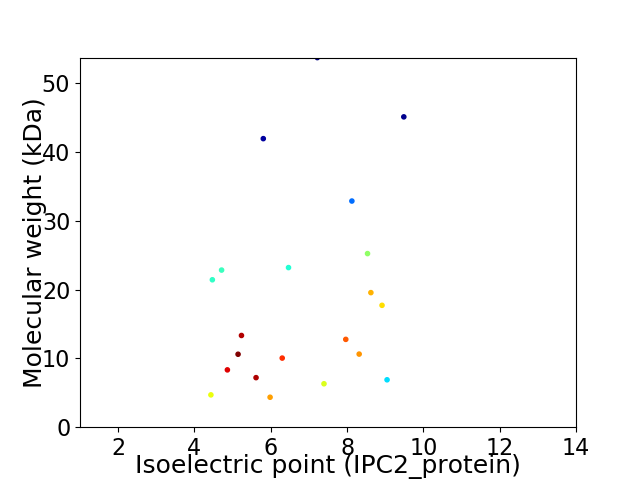
Virtual 2D-PAGE plot for 21 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZNZ1|A0A4D5ZNZ1_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan30 OX=2558619 GN=JavanS30_0017 PE=4 SV=1
MM1 pKa = 7.55RR2 pKa = 11.84TFSDD6 pKa = 3.67TPKK9 pKa = 10.21TFTFHH14 pKa = 5.7YY15 pKa = 8.55TFKK18 pKa = 11.1DD19 pKa = 3.09FDD21 pKa = 4.09TAQVACHH28 pKa = 6.88AILGYY33 pKa = 7.87MTGSYY38 pKa = 7.51EE39 pKa = 4.41QPVIDD44 pKa = 3.87ATYY47 pKa = 10.82HH48 pKa = 6.24NDD50 pKa = 3.59DD51 pKa = 3.72QGGHH55 pKa = 6.59ANQLVLEE62 pKa = 4.2YY63 pKa = 11.33AEE65 pKa = 4.34DD66 pKa = 3.58RR67 pKa = 11.84KK68 pKa = 10.83LNKK71 pKa = 9.29VFKK74 pKa = 10.18RR75 pKa = 11.84ICDD78 pKa = 3.69SFKK81 pKa = 10.93DD82 pKa = 4.21YY83 pKa = 11.52YY84 pKa = 10.51NQPEE88 pKa = 4.31DD89 pKa = 3.62MTDD92 pKa = 3.27EE93 pKa = 4.37EE94 pKa = 5.52LDD96 pKa = 3.85DD97 pKa = 3.95VAQEE101 pKa = 4.22TEE103 pKa = 3.94LLKK106 pKa = 10.77EE107 pKa = 4.24VEE109 pKa = 4.4STDD112 pKa = 3.21KK113 pKa = 11.08KK114 pKa = 10.57RR115 pKa = 11.84VVPLFDD121 pKa = 3.75NSQEE125 pKa = 4.21KK126 pKa = 10.53ADD128 pKa = 3.78KK129 pKa = 10.61QDD131 pKa = 2.52IFMAFISDD139 pKa = 3.71HH140 pKa = 5.87NQLAEE145 pKa = 4.25HH146 pKa = 6.45VSMNYY151 pKa = 10.65KK152 pKa = 10.63KK153 pKa = 8.08MTQEE157 pKa = 3.76DD158 pKa = 3.9LGAVLEE164 pKa = 4.88SISQGFNYY172 pKa = 9.9LYY174 pKa = 11.24DD175 pKa = 3.55MAIEE179 pKa = 4.68GEE181 pKa = 4.45LLVKK185 pKa = 10.71
MM1 pKa = 7.55RR2 pKa = 11.84TFSDD6 pKa = 3.67TPKK9 pKa = 10.21TFTFHH14 pKa = 5.7YY15 pKa = 8.55TFKK18 pKa = 11.1DD19 pKa = 3.09FDD21 pKa = 4.09TAQVACHH28 pKa = 6.88AILGYY33 pKa = 7.87MTGSYY38 pKa = 7.51EE39 pKa = 4.41QPVIDD44 pKa = 3.87ATYY47 pKa = 10.82HH48 pKa = 6.24NDD50 pKa = 3.59DD51 pKa = 3.72QGGHH55 pKa = 6.59ANQLVLEE62 pKa = 4.2YY63 pKa = 11.33AEE65 pKa = 4.34DD66 pKa = 3.58RR67 pKa = 11.84KK68 pKa = 10.83LNKK71 pKa = 9.29VFKK74 pKa = 10.18RR75 pKa = 11.84ICDD78 pKa = 3.69SFKK81 pKa = 10.93DD82 pKa = 4.21YY83 pKa = 11.52YY84 pKa = 10.51NQPEE88 pKa = 4.31DD89 pKa = 3.62MTDD92 pKa = 3.27EE93 pKa = 4.37EE94 pKa = 5.52LDD96 pKa = 3.85DD97 pKa = 3.95VAQEE101 pKa = 4.22TEE103 pKa = 3.94LLKK106 pKa = 10.77EE107 pKa = 4.24VEE109 pKa = 4.4STDD112 pKa = 3.21KK113 pKa = 11.08KK114 pKa = 10.57RR115 pKa = 11.84VVPLFDD121 pKa = 3.75NSQEE125 pKa = 4.21KK126 pKa = 10.53ADD128 pKa = 3.78KK129 pKa = 10.61QDD131 pKa = 2.52IFMAFISDD139 pKa = 3.71HH140 pKa = 5.87NQLAEE145 pKa = 4.25HH146 pKa = 6.45VSMNYY151 pKa = 10.65KK152 pKa = 10.63KK153 pKa = 8.08MTQEE157 pKa = 3.76DD158 pKa = 3.9LGAVLEE164 pKa = 4.88SISQGFNYY172 pKa = 9.9LYY174 pKa = 11.24DD175 pKa = 3.55MAIEE179 pKa = 4.68GEE181 pKa = 4.45LLVKK185 pKa = 10.71
Molecular weight: 21.43 kDa
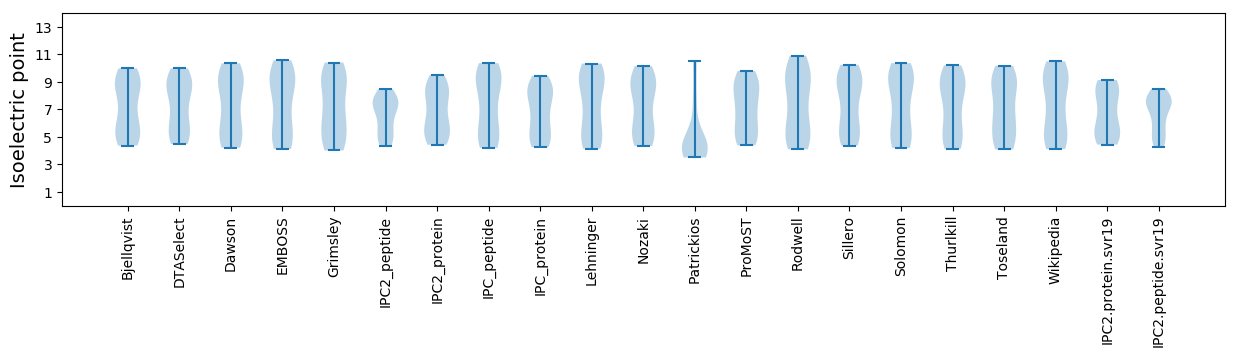
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZNI8|A0A4D5ZNI8_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan30 OX=2558619 GN=JavanS30_0011 PE=4 SV=1
MM1 pKa = 7.85KK2 pKa = 10.27INEE5 pKa = 4.22VKK7 pKa = 10.77KK8 pKa = 10.72KK9 pKa = 10.13DD10 pKa = 3.24GTVIYY15 pKa = 9.86RR16 pKa = 11.84ASVYY20 pKa = 10.73LGTDD24 pKa = 2.51KK25 pKa = 10.85VTGKK29 pKa = 10.64KK30 pKa = 7.91VTTKK34 pKa = 9.18ITGRR38 pKa = 11.84TKK40 pKa = 10.6KK41 pKa = 9.66EE42 pKa = 3.58VRR44 pKa = 11.84EE45 pKa = 3.98KK46 pKa = 10.72AKK48 pKa = 10.48QGAVEE53 pKa = 4.81FIKK56 pKa = 11.1NGSTRR61 pKa = 11.84FKK63 pKa = 9.21ATSVTSYY70 pKa = 11.34QEE72 pKa = 4.91LATLWWDD79 pKa = 3.74SYY81 pKa = 11.1KK82 pKa = 9.85HH83 pKa = 4.21TVKK86 pKa = 10.92YY87 pKa = 8.4NTQLATEE94 pKa = 4.37KK95 pKa = 10.61LLTVHH100 pKa = 6.77VIPIFGAYY108 pKa = 9.53KK109 pKa = 10.15LDD111 pKa = 3.67KK112 pKa = 10.12LTTPLIQSIINKK124 pKa = 9.54LADD127 pKa = 3.45KK128 pKa = 9.75TNKK131 pKa = 10.06GEE133 pKa = 4.09RR134 pKa = 11.84KK135 pKa = 9.79AYY137 pKa = 9.6LHH139 pKa = 6.59YY140 pKa = 11.09DD141 pKa = 4.1RR142 pKa = 11.84IHH144 pKa = 6.67ALNKK148 pKa = 10.16RR149 pKa = 11.84ILQYY153 pKa = 10.96GVIMQAIPFNPARR166 pKa = 11.84EE167 pKa = 4.24VILPRR172 pKa = 11.84NTKK175 pKa = 9.3KK176 pKa = 10.99ANTKK180 pKa = 9.27RR181 pKa = 11.84VKK183 pKa = 10.5HH184 pKa = 5.63FEE186 pKa = 3.77NDD188 pKa = 3.02EE189 pKa = 3.81LRR191 pKa = 11.84TFFNYY196 pKa = 10.41LNNLDD201 pKa = 3.4KK202 pKa = 11.07SKK204 pKa = 10.89YY205 pKa = 7.95RR206 pKa = 11.84NFYY209 pKa = 10.5EE210 pKa = 3.92VTLYY214 pKa = 11.03KK215 pKa = 10.48FLLATGCRR223 pKa = 11.84INEE226 pKa = 3.8ALALNWSDD234 pKa = 3.65IDD236 pKa = 3.87LDD238 pKa = 3.8NAVVHH243 pKa = 5.5ITKK246 pKa = 9.24TLNYY250 pKa = 8.48KK251 pKa = 9.4QEE253 pKa = 4.1INSPKK258 pKa = 10.23SKK260 pKa = 10.48SSYY263 pKa = 10.17RR264 pKa = 11.84DD265 pKa = 2.84IDD267 pKa = 3.51IDD269 pKa = 3.84SQTITMLKK277 pKa = 9.08QYY279 pKa = 10.12RR280 pKa = 11.84RR281 pKa = 11.84RR282 pKa = 11.84QIQEE286 pKa = 2.96AWKK289 pKa = 9.76LGRR292 pKa = 11.84SEE294 pKa = 4.29TVVFSDD300 pKa = 6.41FIHH303 pKa = 7.11KK304 pKa = 10.24YY305 pKa = 8.57PNNRR309 pKa = 11.84TLQTRR314 pKa = 11.84LRR316 pKa = 11.84THH318 pKa = 6.95FKK320 pKa = 10.18RR321 pKa = 11.84ANVPNIGFHH330 pKa = 6.08GFRR333 pKa = 11.84HH334 pKa = 4.87THH336 pKa = 6.65ASLLLNTGIPYY347 pKa = 10.09KK348 pKa = 10.14EE349 pKa = 3.64LQYY352 pKa = 11.41RR353 pKa = 11.84LGHH356 pKa = 5.67STLSMTMDD364 pKa = 3.58IYY366 pKa = 11.68SHH368 pKa = 6.98LSKK371 pKa = 11.0EE372 pKa = 4.25NAKK375 pKa = 10.34KK376 pKa = 10.33AVSFFEE382 pKa = 4.08TAINSII388 pKa = 4.07
MM1 pKa = 7.85KK2 pKa = 10.27INEE5 pKa = 4.22VKK7 pKa = 10.77KK8 pKa = 10.72KK9 pKa = 10.13DD10 pKa = 3.24GTVIYY15 pKa = 9.86RR16 pKa = 11.84ASVYY20 pKa = 10.73LGTDD24 pKa = 2.51KK25 pKa = 10.85VTGKK29 pKa = 10.64KK30 pKa = 7.91VTTKK34 pKa = 9.18ITGRR38 pKa = 11.84TKK40 pKa = 10.6KK41 pKa = 9.66EE42 pKa = 3.58VRR44 pKa = 11.84EE45 pKa = 3.98KK46 pKa = 10.72AKK48 pKa = 10.48QGAVEE53 pKa = 4.81FIKK56 pKa = 11.1NGSTRR61 pKa = 11.84FKK63 pKa = 9.21ATSVTSYY70 pKa = 11.34QEE72 pKa = 4.91LATLWWDD79 pKa = 3.74SYY81 pKa = 11.1KK82 pKa = 9.85HH83 pKa = 4.21TVKK86 pKa = 10.92YY87 pKa = 8.4NTQLATEE94 pKa = 4.37KK95 pKa = 10.61LLTVHH100 pKa = 6.77VIPIFGAYY108 pKa = 9.53KK109 pKa = 10.15LDD111 pKa = 3.67KK112 pKa = 10.12LTTPLIQSIINKK124 pKa = 9.54LADD127 pKa = 3.45KK128 pKa = 9.75TNKK131 pKa = 10.06GEE133 pKa = 4.09RR134 pKa = 11.84KK135 pKa = 9.79AYY137 pKa = 9.6LHH139 pKa = 6.59YY140 pKa = 11.09DD141 pKa = 4.1RR142 pKa = 11.84IHH144 pKa = 6.67ALNKK148 pKa = 10.16RR149 pKa = 11.84ILQYY153 pKa = 10.96GVIMQAIPFNPARR166 pKa = 11.84EE167 pKa = 4.24VILPRR172 pKa = 11.84NTKK175 pKa = 9.3KK176 pKa = 10.99ANTKK180 pKa = 9.27RR181 pKa = 11.84VKK183 pKa = 10.5HH184 pKa = 5.63FEE186 pKa = 3.77NDD188 pKa = 3.02EE189 pKa = 3.81LRR191 pKa = 11.84TFFNYY196 pKa = 10.41LNNLDD201 pKa = 3.4KK202 pKa = 11.07SKK204 pKa = 10.89YY205 pKa = 7.95RR206 pKa = 11.84NFYY209 pKa = 10.5EE210 pKa = 3.92VTLYY214 pKa = 11.03KK215 pKa = 10.48FLLATGCRR223 pKa = 11.84INEE226 pKa = 3.8ALALNWSDD234 pKa = 3.65IDD236 pKa = 3.87LDD238 pKa = 3.8NAVVHH243 pKa = 5.5ITKK246 pKa = 9.24TLNYY250 pKa = 8.48KK251 pKa = 9.4QEE253 pKa = 4.1INSPKK258 pKa = 10.23SKK260 pKa = 10.48SSYY263 pKa = 10.17RR264 pKa = 11.84DD265 pKa = 2.84IDD267 pKa = 3.51IDD269 pKa = 3.84SQTITMLKK277 pKa = 9.08QYY279 pKa = 10.12RR280 pKa = 11.84RR281 pKa = 11.84RR282 pKa = 11.84QIQEE286 pKa = 2.96AWKK289 pKa = 9.76LGRR292 pKa = 11.84SEE294 pKa = 4.29TVVFSDD300 pKa = 6.41FIHH303 pKa = 7.11KK304 pKa = 10.24YY305 pKa = 8.57PNNRR309 pKa = 11.84TLQTRR314 pKa = 11.84LRR316 pKa = 11.84THH318 pKa = 6.95FKK320 pKa = 10.18RR321 pKa = 11.84ANVPNIGFHH330 pKa = 6.08GFRR333 pKa = 11.84HH334 pKa = 4.87THH336 pKa = 6.65ASLLLNTGIPYY347 pKa = 10.09KK348 pKa = 10.14EE349 pKa = 3.64LQYY352 pKa = 11.41RR353 pKa = 11.84LGHH356 pKa = 5.67STLSMTMDD364 pKa = 3.58IYY366 pKa = 11.68SHH368 pKa = 6.98LSKK371 pKa = 11.0EE372 pKa = 4.25NAKK375 pKa = 10.34KK376 pKa = 10.33AVSFFEE382 pKa = 4.08TAINSII388 pKa = 4.07
Molecular weight: 45.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3454 |
39 |
464 |
164.5 |
18.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.877 ± 0.417 | 0.811 ± 0.256 |
6.109 ± 0.508 | 7.759 ± 0.548 |
4.082 ± 0.294 | 4.748 ± 0.244 |
1.94 ± 0.303 | 6.369 ± 0.404 |
9.554 ± 0.433 | 8.715 ± 0.537 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.142 ± 0.372 | 5.414 ± 0.377 |
3.098 ± 0.493 | 4.574 ± 0.285 |
4.922 ± 0.436 | 6.138 ± 0.859 |
6.717 ± 0.488 | 5.124 ± 0.406 |
0.898 ± 0.19 | 5.009 ± 0.383 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
