
Cardamine chlorotic fleck virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Betacarmovirus
Average proteome isoelectric point is 8.56
Get precalculated fractions of proteins
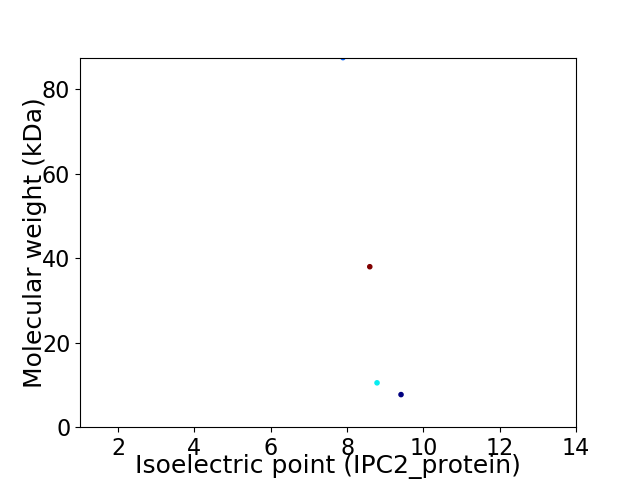
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q65988|Q65988_9TOMB p8 protein OS=Cardamine chlorotic fleck virus OX=31711 PE=4 SV=1
MM1 pKa = 8.06DD2 pKa = 5.48ALLPEE7 pKa = 4.61FLAHH11 pKa = 6.81KK12 pKa = 10.38LSRR15 pKa = 11.84LVLKK19 pKa = 10.67CFGIGSDD26 pKa = 4.09EE27 pKa = 4.34EE28 pKa = 4.32APQQIFVGEE37 pKa = 3.96FPVWGGEE44 pKa = 3.97IVPPQPEE51 pKa = 4.36EE52 pKa = 3.99LNQLVIPRR60 pKa = 11.84KK61 pKa = 6.81WVRR64 pKa = 11.84SIGSSLNRR72 pKa = 11.84VVLPPSSGVVASLEE86 pKa = 4.34EE87 pKa = 3.98NCVAPFEE94 pKa = 4.34EE95 pKa = 4.97ASVEE99 pKa = 4.06QLEE102 pKa = 4.42PASGDD107 pKa = 3.38TGDD110 pKa = 3.57LPRR113 pKa = 11.84RR114 pKa = 11.84KK115 pKa = 9.0IRR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84GRR121 pKa = 11.84FVAHH125 pKa = 6.53AVCAAKK131 pKa = 10.43LHH133 pKa = 6.26FSGCPTQTLANRR145 pKa = 11.84LAVSKK150 pKa = 10.16WLVQYY155 pKa = 8.95CTEE158 pKa = 4.12RR159 pKa = 11.84NVIPSHH165 pKa = 5.02TRR167 pKa = 11.84IAVQEE172 pKa = 4.02ALPQVFLPDD181 pKa = 3.22GHH183 pKa = 6.59DD184 pKa = 3.41LKK186 pKa = 11.16EE187 pKa = 4.09VLDD190 pKa = 3.83LHH192 pKa = 6.03TEE194 pKa = 4.01EE195 pKa = 4.63AHH197 pKa = 6.99DD198 pKa = 4.18RR199 pKa = 11.84RR200 pKa = 11.84NALRR204 pKa = 11.84KK205 pKa = 9.25RR206 pKa = 11.84GRR208 pKa = 11.84HH209 pKa = 3.67VRR211 pKa = 11.84RR212 pKa = 11.84WWVQLAMHH220 pKa = 7.12PMTGRR225 pKa = 11.84SWLRR229 pKa = 11.84AWRR232 pKa = 11.84RR233 pKa = 11.84LTGLPDD239 pKa = 3.56DD240 pKa = 4.17VAVQFVRR247 pKa = 11.84XGCLRR252 pKa = 11.84EE253 pKa = 3.9LVGRR257 pKa = 11.84EE258 pKa = 3.66TAISRR263 pKa = 11.84GEE265 pKa = 4.03HH266 pKa = 5.33PAMRR270 pKa = 11.84VFPLAQPPKK279 pKa = 9.18IRR281 pKa = 11.84RR282 pKa = 11.84IFHH285 pKa = 5.58ICGMGNGLDD294 pKa = 3.67FGVHH298 pKa = 5.81NNSLNNLRR306 pKa = 11.84RR307 pKa = 11.84GLMEE311 pKa = 3.52RR312 pKa = 11.84VFYY315 pKa = 11.45VEE317 pKa = 5.05DD318 pKa = 3.59AQKK321 pKa = 10.38EE322 pKa = 4.38LKK324 pKa = 9.72PAPQPLPDD332 pKa = 4.31AFAKK336 pKa = 10.7LSGIRR341 pKa = 11.84RR342 pKa = 11.84KK343 pKa = 9.47LVKK346 pKa = 10.25LAGNHH351 pKa = 4.65TPILRR356 pKa = 11.84EE357 pKa = 4.57DD358 pKa = 3.54YY359 pKa = 10.9SKK361 pKa = 11.1LYY363 pKa = 9.71KK364 pKa = 10.26GRR366 pKa = 11.84RR367 pKa = 11.84ATIYY371 pKa = 10.77QNACEE376 pKa = 4.19SLADD380 pKa = 4.1RR381 pKa = 11.84ACTQGGSAEE390 pKa = 4.24LKK392 pKa = 9.4TFVKK396 pKa = 10.54AEE398 pKa = 4.43KK399 pKa = 10.48INFTAKK405 pKa = 10.05KK406 pKa = 9.71DD407 pKa = 3.4PAPRR411 pKa = 11.84VIQPRR416 pKa = 11.84DD417 pKa = 3.13PRR419 pKa = 11.84YY420 pKa = 9.34NIEE423 pKa = 3.55VGKK426 pKa = 9.76YY427 pKa = 7.56LKK429 pKa = 10.49CYY431 pKa = 7.6EE432 pKa = 4.09HH433 pKa = 6.8HH434 pKa = 7.45LYY436 pKa = 10.48RR437 pKa = 11.84AIDD440 pKa = 3.78KK441 pKa = 7.23MWCGPTVLKK450 pKa = 10.52GYY452 pKa = 9.78NVEE455 pKa = 4.24EE456 pKa = 4.49IGNIMSDD463 pKa = 3.12AWNQFQKK470 pKa = 10.62PCAIGFDD477 pKa = 3.65MKK479 pKa = 10.85RR480 pKa = 11.84FDD482 pKa = 3.31QHH484 pKa = 8.09VSVDD488 pKa = 3.44ALKK491 pKa = 10.34WEE493 pKa = 4.08HH494 pKa = 5.97SVYY497 pKa = 10.36NASFNCPDD505 pKa = 3.86LAKK508 pKa = 10.26MLTWQLKK515 pKa = 8.38NKK517 pKa = 9.91GVGRR521 pKa = 11.84ASDD524 pKa = 4.59GYY526 pKa = 10.41IKK528 pKa = 10.74YY529 pKa = 9.75QVEE532 pKa = 4.47GCRR535 pKa = 11.84MSGDD539 pKa = 3.46VNTAMGNCLLACSITRR555 pKa = 11.84YY556 pKa = 9.66LMKK559 pKa = 10.5GIKK562 pKa = 9.5SRR564 pKa = 11.84LINNGDD570 pKa = 3.48DD571 pKa = 3.1CVVFFEE577 pKa = 4.71SGDD580 pKa = 3.34MHH582 pKa = 6.32EE583 pKa = 4.33VRR585 pKa = 11.84EE586 pKa = 4.26RR587 pKa = 11.84LCRR590 pKa = 11.84WIDD593 pKa = 3.6FGFQCIAEE601 pKa = 4.21EE602 pKa = 3.92PQYY605 pKa = 11.33DD606 pKa = 3.84LEE608 pKa = 4.55KK609 pKa = 11.26VEE611 pKa = 4.75FCQMSPIYY619 pKa = 10.43DD620 pKa = 3.55GEE622 pKa = 4.29GWVMVRR628 pKa = 11.84NPRR631 pKa = 11.84VSLSKK636 pKa = 10.67DD637 pKa = 3.38SYY639 pKa = 11.35STTQWGSVRR648 pKa = 11.84DD649 pKa = 3.74AASWLASIGEE659 pKa = 4.41CGMAIAGGIPIVQSYY674 pKa = 7.76YY675 pKa = 11.09NCLLRR680 pKa = 11.84NFKK683 pKa = 10.27HH684 pKa = 6.57LARR687 pKa = 11.84NTSKK691 pKa = 10.98KK692 pKa = 10.49KK693 pKa = 10.63LDD695 pKa = 3.31VSFNSGFSMLAQNGNRR711 pKa = 11.84GSKK714 pKa = 9.49EE715 pKa = 3.57VSEE718 pKa = 4.19AARR721 pKa = 11.84YY722 pKa = 9.0SFYY725 pKa = 11.33VAFGYY730 pKa = 8.92TPDD733 pKa = 3.61EE734 pKa = 3.95QRR736 pKa = 11.84GLEE739 pKa = 4.1GYY741 pKa = 10.08YY742 pKa = 10.99DD743 pKa = 3.86NLALEE748 pKa = 4.72AAWDD752 pKa = 3.77PEE754 pKa = 4.64GYY756 pKa = 10.46DD757 pKa = 3.35EE758 pKa = 4.58TLLNRR763 pKa = 11.84WIINPSLTTPP773 pKa = 3.72
MM1 pKa = 8.06DD2 pKa = 5.48ALLPEE7 pKa = 4.61FLAHH11 pKa = 6.81KK12 pKa = 10.38LSRR15 pKa = 11.84LVLKK19 pKa = 10.67CFGIGSDD26 pKa = 4.09EE27 pKa = 4.34EE28 pKa = 4.32APQQIFVGEE37 pKa = 3.96FPVWGGEE44 pKa = 3.97IVPPQPEE51 pKa = 4.36EE52 pKa = 3.99LNQLVIPRR60 pKa = 11.84KK61 pKa = 6.81WVRR64 pKa = 11.84SIGSSLNRR72 pKa = 11.84VVLPPSSGVVASLEE86 pKa = 4.34EE87 pKa = 3.98NCVAPFEE94 pKa = 4.34EE95 pKa = 4.97ASVEE99 pKa = 4.06QLEE102 pKa = 4.42PASGDD107 pKa = 3.38TGDD110 pKa = 3.57LPRR113 pKa = 11.84RR114 pKa = 11.84KK115 pKa = 9.0IRR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84GRR121 pKa = 11.84FVAHH125 pKa = 6.53AVCAAKK131 pKa = 10.43LHH133 pKa = 6.26FSGCPTQTLANRR145 pKa = 11.84LAVSKK150 pKa = 10.16WLVQYY155 pKa = 8.95CTEE158 pKa = 4.12RR159 pKa = 11.84NVIPSHH165 pKa = 5.02TRR167 pKa = 11.84IAVQEE172 pKa = 4.02ALPQVFLPDD181 pKa = 3.22GHH183 pKa = 6.59DD184 pKa = 3.41LKK186 pKa = 11.16EE187 pKa = 4.09VLDD190 pKa = 3.83LHH192 pKa = 6.03TEE194 pKa = 4.01EE195 pKa = 4.63AHH197 pKa = 6.99DD198 pKa = 4.18RR199 pKa = 11.84RR200 pKa = 11.84NALRR204 pKa = 11.84KK205 pKa = 9.25RR206 pKa = 11.84GRR208 pKa = 11.84HH209 pKa = 3.67VRR211 pKa = 11.84RR212 pKa = 11.84WWVQLAMHH220 pKa = 7.12PMTGRR225 pKa = 11.84SWLRR229 pKa = 11.84AWRR232 pKa = 11.84RR233 pKa = 11.84LTGLPDD239 pKa = 3.56DD240 pKa = 4.17VAVQFVRR247 pKa = 11.84XGCLRR252 pKa = 11.84EE253 pKa = 3.9LVGRR257 pKa = 11.84EE258 pKa = 3.66TAISRR263 pKa = 11.84GEE265 pKa = 4.03HH266 pKa = 5.33PAMRR270 pKa = 11.84VFPLAQPPKK279 pKa = 9.18IRR281 pKa = 11.84RR282 pKa = 11.84IFHH285 pKa = 5.58ICGMGNGLDD294 pKa = 3.67FGVHH298 pKa = 5.81NNSLNNLRR306 pKa = 11.84RR307 pKa = 11.84GLMEE311 pKa = 3.52RR312 pKa = 11.84VFYY315 pKa = 11.45VEE317 pKa = 5.05DD318 pKa = 3.59AQKK321 pKa = 10.38EE322 pKa = 4.38LKK324 pKa = 9.72PAPQPLPDD332 pKa = 4.31AFAKK336 pKa = 10.7LSGIRR341 pKa = 11.84RR342 pKa = 11.84KK343 pKa = 9.47LVKK346 pKa = 10.25LAGNHH351 pKa = 4.65TPILRR356 pKa = 11.84EE357 pKa = 4.57DD358 pKa = 3.54YY359 pKa = 10.9SKK361 pKa = 11.1LYY363 pKa = 9.71KK364 pKa = 10.26GRR366 pKa = 11.84RR367 pKa = 11.84ATIYY371 pKa = 10.77QNACEE376 pKa = 4.19SLADD380 pKa = 4.1RR381 pKa = 11.84ACTQGGSAEE390 pKa = 4.24LKK392 pKa = 9.4TFVKK396 pKa = 10.54AEE398 pKa = 4.43KK399 pKa = 10.48INFTAKK405 pKa = 10.05KK406 pKa = 9.71DD407 pKa = 3.4PAPRR411 pKa = 11.84VIQPRR416 pKa = 11.84DD417 pKa = 3.13PRR419 pKa = 11.84YY420 pKa = 9.34NIEE423 pKa = 3.55VGKK426 pKa = 9.76YY427 pKa = 7.56LKK429 pKa = 10.49CYY431 pKa = 7.6EE432 pKa = 4.09HH433 pKa = 6.8HH434 pKa = 7.45LYY436 pKa = 10.48RR437 pKa = 11.84AIDD440 pKa = 3.78KK441 pKa = 7.23MWCGPTVLKK450 pKa = 10.52GYY452 pKa = 9.78NVEE455 pKa = 4.24EE456 pKa = 4.49IGNIMSDD463 pKa = 3.12AWNQFQKK470 pKa = 10.62PCAIGFDD477 pKa = 3.65MKK479 pKa = 10.85RR480 pKa = 11.84FDD482 pKa = 3.31QHH484 pKa = 8.09VSVDD488 pKa = 3.44ALKK491 pKa = 10.34WEE493 pKa = 4.08HH494 pKa = 5.97SVYY497 pKa = 10.36NASFNCPDD505 pKa = 3.86LAKK508 pKa = 10.26MLTWQLKK515 pKa = 8.38NKK517 pKa = 9.91GVGRR521 pKa = 11.84ASDD524 pKa = 4.59GYY526 pKa = 10.41IKK528 pKa = 10.74YY529 pKa = 9.75QVEE532 pKa = 4.47GCRR535 pKa = 11.84MSGDD539 pKa = 3.46VNTAMGNCLLACSITRR555 pKa = 11.84YY556 pKa = 9.66LMKK559 pKa = 10.5GIKK562 pKa = 9.5SRR564 pKa = 11.84LINNGDD570 pKa = 3.48DD571 pKa = 3.1CVVFFEE577 pKa = 4.71SGDD580 pKa = 3.34MHH582 pKa = 6.32EE583 pKa = 4.33VRR585 pKa = 11.84EE586 pKa = 4.26RR587 pKa = 11.84LCRR590 pKa = 11.84WIDD593 pKa = 3.6FGFQCIAEE601 pKa = 4.21EE602 pKa = 3.92PQYY605 pKa = 11.33DD606 pKa = 3.84LEE608 pKa = 4.55KK609 pKa = 11.26VEE611 pKa = 4.75FCQMSPIYY619 pKa = 10.43DD620 pKa = 3.55GEE622 pKa = 4.29GWVMVRR628 pKa = 11.84NPRR631 pKa = 11.84VSLSKK636 pKa = 10.67DD637 pKa = 3.38SYY639 pKa = 11.35STTQWGSVRR648 pKa = 11.84DD649 pKa = 3.74AASWLASIGEE659 pKa = 4.41CGMAIAGGIPIVQSYY674 pKa = 7.76YY675 pKa = 11.09NCLLRR680 pKa = 11.84NFKK683 pKa = 10.27HH684 pKa = 6.57LARR687 pKa = 11.84NTSKK691 pKa = 10.98KK692 pKa = 10.49KK693 pKa = 10.63LDD695 pKa = 3.31VSFNSGFSMLAQNGNRR711 pKa = 11.84GSKK714 pKa = 9.49EE715 pKa = 3.57VSEE718 pKa = 4.19AARR721 pKa = 11.84YY722 pKa = 9.0SFYY725 pKa = 11.33VAFGYY730 pKa = 8.92TPDD733 pKa = 3.61EE734 pKa = 3.95QRR736 pKa = 11.84GLEE739 pKa = 4.1GYY741 pKa = 10.08YY742 pKa = 10.99DD743 pKa = 3.86NLALEE748 pKa = 4.72AAWDD752 pKa = 3.77PEE754 pKa = 4.64GYY756 pKa = 10.46DD757 pKa = 3.35EE758 pKa = 4.58TLLNRR763 pKa = 11.84WIINPSLTTPP773 pKa = 3.72
Molecular weight: 87.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q65989|Q65989_9TOMB p9 protein OS=Cardamine chlorotic fleck virus OX=31711 PE=4 SV=1
MM1 pKa = 8.11DD2 pKa = 4.4NQSEE6 pKa = 4.4SDD8 pKa = 3.39NSVKK12 pKa = 10.16RR13 pKa = 11.84RR14 pKa = 11.84SGKK17 pKa = 9.81KK18 pKa = 8.45VNKK21 pKa = 8.25QVRR24 pKa = 11.84GSEE27 pKa = 3.72KK28 pKa = 10.1SATKK32 pKa = 10.37RR33 pKa = 11.84SVASHH38 pKa = 6.93AARR41 pKa = 11.84SFSQRR46 pKa = 11.84PSGDD50 pKa = 3.3SAGGNWVLVADD61 pKa = 5.01KK62 pKa = 10.73IEE64 pKa = 3.97VSISFNFF71 pKa = 3.34
MM1 pKa = 8.11DD2 pKa = 4.4NQSEE6 pKa = 4.4SDD8 pKa = 3.39NSVKK12 pKa = 10.16RR13 pKa = 11.84RR14 pKa = 11.84SGKK17 pKa = 9.81KK18 pKa = 8.45VNKK21 pKa = 8.25QVRR24 pKa = 11.84GSEE27 pKa = 3.72KK28 pKa = 10.1SATKK32 pKa = 10.37RR33 pKa = 11.84SVASHH38 pKa = 6.93AARR41 pKa = 11.84SFSQRR46 pKa = 11.84PSGDD50 pKa = 3.3SAGGNWVLVADD61 pKa = 5.01KK62 pKa = 10.73IEE64 pKa = 3.97VSISFNFF71 pKa = 3.34
Molecular weight: 7.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1289 |
71 |
773 |
322.3 |
35.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.991 ± 0.633 | 2.095 ± 0.71 |
4.577 ± 0.348 | 5.586 ± 1.102 |
3.801 ± 0.105 | 7.292 ± 0.694 |
2.095 ± 0.424 | 4.577 ± 0.758 |
5.818 ± 0.669 | 8.999 ± 1.264 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.784 ± 0.494 | 4.422 ± 0.633 |
5.431 ± 0.494 | 3.646 ± 0.214 |
6.827 ± 1.064 | 7.758 ± 2.255 |
4.81 ± 1.653 | 7.448 ± 0.521 |
2.017 ± 0.18 | 2.948 ± 0.441 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
