
Rhizophagus sp. RF1 mitovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
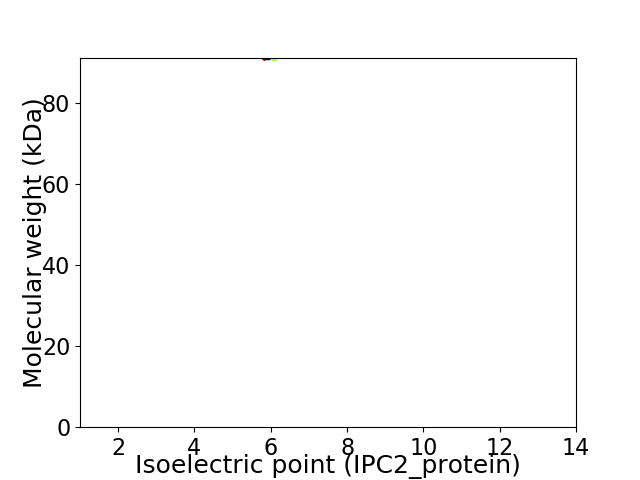
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E2RWR3|E2RWR3_9VIRU RNA-directed RNA polymerase OS=Rhizophagus sp. RF1 mitovirus OX=758866 GN=RdRp PE=4 SV=2
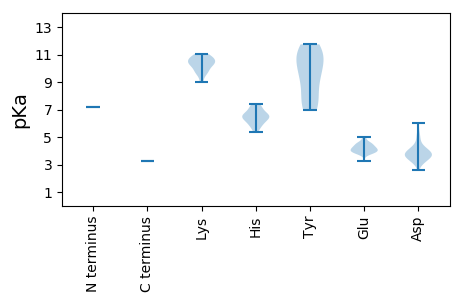
MM1 pKa = 7.15STFNFNSPNEE11 pKa = 4.04VRR13 pKa = 11.84RR14 pKa = 11.84VTRR17 pKa = 11.84SNIASLFAALMHH29 pKa = 6.55LNSLVTVTYY38 pKa = 9.13SQRR41 pKa = 11.84LLAFLGAVYY50 pKa = 10.82NRR52 pKa = 11.84ILVLFDD58 pKa = 3.76ANARR62 pKa = 11.84SLISEE67 pKa = 4.59LKK69 pKa = 10.32LVRR72 pKa = 11.84RR73 pKa = 11.84WFLEE77 pKa = 4.45FIRR80 pKa = 11.84NGNTDD85 pKa = 3.39NPGLEE90 pKa = 3.86WDD92 pKa = 4.1RR93 pKa = 11.84WDD95 pKa = 5.14DD96 pKa = 4.2SNNCPVLLEE105 pKa = 4.52GLDD108 pKa = 3.85ALWDD112 pKa = 4.55EE113 pKa = 4.94IEE115 pKa = 4.56SPSDD119 pKa = 3.12SHH121 pKa = 5.98YY122 pKa = 9.92TDD124 pKa = 4.25HH125 pKa = 7.42AAQLIFTLLSIDD137 pKa = 4.15RR138 pKa = 11.84IIVVPAVPNYY148 pKa = 9.48STIEE152 pKa = 4.35DD153 pKa = 4.19GPVIEE158 pKa = 4.36PVGYY162 pKa = 6.95PTNEE166 pKa = 3.85EE167 pKa = 4.02LVNALSSLHH176 pKa = 6.74IDD178 pKa = 3.4PAAFKK183 pKa = 10.73AFYY186 pKa = 9.08NQQVHH191 pKa = 6.66DD192 pKa = 4.22FDD194 pKa = 5.39YY195 pKa = 11.22EE196 pKa = 4.3VLSTRR201 pKa = 11.84GPNGDD206 pKa = 3.94ATWTAHH212 pKa = 6.22LDD214 pKa = 3.31ARR216 pKa = 11.84AWALDD221 pKa = 3.42TEE223 pKa = 4.5LFRR226 pKa = 11.84RR227 pKa = 11.84FSAWLEE233 pKa = 3.76EE234 pKa = 4.16SRR236 pKa = 11.84LTRR239 pKa = 11.84ILRR242 pKa = 11.84DD243 pKa = 3.17LFGCIRR249 pKa = 11.84SAAAEE254 pKa = 4.66AIPNLSPILGKK265 pKa = 10.73LSVIEE270 pKa = 4.01EE271 pKa = 4.27WGGKK275 pKa = 9.01ARR277 pKa = 11.84IVAQMDD283 pKa = 3.68YY284 pKa = 7.44WTQMALTPLHH294 pKa = 5.34NTINHH299 pKa = 6.63FLRR302 pKa = 11.84ALKK305 pKa = 10.43EE306 pKa = 4.21DD307 pKa = 3.65GTFNQHH313 pKa = 7.04AIAEE317 pKa = 4.48RR318 pKa = 11.84VRR320 pKa = 11.84QWTADD325 pKa = 3.04PSMEE329 pKa = 4.14VFSFDD334 pKa = 3.07LTAATDD340 pKa = 3.63RR341 pKa = 11.84VPITFQEE348 pKa = 4.57SILSYY353 pKa = 11.4LMTSKK358 pKa = 10.86SFGNGWASILVDD370 pKa = 4.94RR371 pKa = 11.84EE372 pKa = 3.87FLTPNGDD379 pKa = 3.92LISYY383 pKa = 7.6NTGQPMGARR392 pKa = 11.84SSFPMLALTHH402 pKa = 6.48HH403 pKa = 7.23IIVQIAAARR412 pKa = 11.84AGLTVYY418 pKa = 10.44RR419 pKa = 11.84DD420 pKa = 3.73YY421 pKa = 11.74VVLGDD426 pKa = 6.02DD427 pKa = 3.61VTLTNAQVAAHH438 pKa = 6.11YY439 pKa = 7.21QTIMRR444 pKa = 11.84CLGVPINLSKK454 pKa = 11.05SIVHH458 pKa = 5.82VDD460 pKa = 3.32GGVSMAEE467 pKa = 3.23ICKK470 pKa = 9.85RR471 pKa = 11.84VFMDD475 pKa = 3.91GVEE478 pKa = 3.91ISRR481 pKa = 11.84FNPKK485 pKa = 10.33LIVNVIRR492 pKa = 11.84DD493 pKa = 3.74GRR495 pKa = 11.84LGPDD499 pKa = 3.7LQNDD503 pKa = 4.77LIIRR507 pKa = 11.84GWDD510 pKa = 3.29PSNEE514 pKa = 4.14VFWKK518 pKa = 10.25FMAGLLSIDD527 pKa = 4.01NLTLLIRR534 pKa = 11.84LNCAPISITGLLRR547 pKa = 11.84QFASNSKK554 pKa = 9.73LAQLSAWIPAYY565 pKa = 10.19QDD567 pKa = 3.69LKK569 pKa = 10.82PEE571 pKa = 4.19HH572 pKa = 6.55LVEE575 pKa = 4.29LFTYY579 pKa = 8.26VTASEE584 pKa = 3.96ALKK587 pKa = 10.76RR588 pKa = 11.84LDD590 pKa = 4.43GILRR594 pKa = 11.84AAVTINDD601 pKa = 3.94SLSIIAAANAHH612 pKa = 7.13PDD614 pKa = 4.05RR615 pKa = 11.84IPQYY619 pKa = 11.31VRR621 pKa = 11.84DD622 pKa = 3.61TWLGEE627 pKa = 3.86GLTKK631 pKa = 10.28EE632 pKa = 3.75EE633 pKa = 4.12RR634 pKa = 11.84ARR636 pKa = 11.84LEE638 pKa = 3.79GLIASTGPITPNHH651 pKa = 6.42PLVSASRR658 pKa = 11.84AEE660 pKa = 3.93ANRR663 pKa = 11.84ISEE666 pKa = 4.98LLHH669 pKa = 5.75QLNSHH674 pKa = 6.45DD675 pKa = 3.57TAIITRR681 pKa = 11.84ARR683 pKa = 11.84LGLLDD688 pKa = 3.58VFRR691 pKa = 11.84TSISSIWLDD700 pKa = 3.82DD701 pKa = 3.82GNIRR705 pKa = 11.84AGEE708 pKa = 4.02SRR710 pKa = 11.84SIFTRR715 pKa = 11.84MLTTLVSLFTSEE727 pKa = 4.23KK728 pKa = 9.76RR729 pKa = 11.84VSKK732 pKa = 10.52SGRR735 pKa = 11.84NLSLSYY741 pKa = 10.88SVVLTSLSRR750 pKa = 11.84LWTVALDD757 pKa = 3.61FGGQVTVNALRR768 pKa = 11.84ANVTRR773 pKa = 11.84DD774 pKa = 2.63IHH776 pKa = 6.35NAVDD780 pKa = 3.5NLKK783 pKa = 10.35AAEE786 pKa = 4.0EE787 pKa = 4.01AAVLISSSSVPTTSPTPATPKK808 pKa = 9.6GIRR811 pKa = 11.84RR812 pKa = 11.84RR813 pKa = 11.84RR814 pKa = 11.84AFARR818 pKa = 11.84ISS820 pKa = 3.22
MM1 pKa = 7.15STFNFNSPNEE11 pKa = 4.04VRR13 pKa = 11.84RR14 pKa = 11.84VTRR17 pKa = 11.84SNIASLFAALMHH29 pKa = 6.55LNSLVTVTYY38 pKa = 9.13SQRR41 pKa = 11.84LLAFLGAVYY50 pKa = 10.82NRR52 pKa = 11.84ILVLFDD58 pKa = 3.76ANARR62 pKa = 11.84SLISEE67 pKa = 4.59LKK69 pKa = 10.32LVRR72 pKa = 11.84RR73 pKa = 11.84WFLEE77 pKa = 4.45FIRR80 pKa = 11.84NGNTDD85 pKa = 3.39NPGLEE90 pKa = 3.86WDD92 pKa = 4.1RR93 pKa = 11.84WDD95 pKa = 5.14DD96 pKa = 4.2SNNCPVLLEE105 pKa = 4.52GLDD108 pKa = 3.85ALWDD112 pKa = 4.55EE113 pKa = 4.94IEE115 pKa = 4.56SPSDD119 pKa = 3.12SHH121 pKa = 5.98YY122 pKa = 9.92TDD124 pKa = 4.25HH125 pKa = 7.42AAQLIFTLLSIDD137 pKa = 4.15RR138 pKa = 11.84IIVVPAVPNYY148 pKa = 9.48STIEE152 pKa = 4.35DD153 pKa = 4.19GPVIEE158 pKa = 4.36PVGYY162 pKa = 6.95PTNEE166 pKa = 3.85EE167 pKa = 4.02LVNALSSLHH176 pKa = 6.74IDD178 pKa = 3.4PAAFKK183 pKa = 10.73AFYY186 pKa = 9.08NQQVHH191 pKa = 6.66DD192 pKa = 4.22FDD194 pKa = 5.39YY195 pKa = 11.22EE196 pKa = 4.3VLSTRR201 pKa = 11.84GPNGDD206 pKa = 3.94ATWTAHH212 pKa = 6.22LDD214 pKa = 3.31ARR216 pKa = 11.84AWALDD221 pKa = 3.42TEE223 pKa = 4.5LFRR226 pKa = 11.84RR227 pKa = 11.84FSAWLEE233 pKa = 3.76EE234 pKa = 4.16SRR236 pKa = 11.84LTRR239 pKa = 11.84ILRR242 pKa = 11.84DD243 pKa = 3.17LFGCIRR249 pKa = 11.84SAAAEE254 pKa = 4.66AIPNLSPILGKK265 pKa = 10.73LSVIEE270 pKa = 4.01EE271 pKa = 4.27WGGKK275 pKa = 9.01ARR277 pKa = 11.84IVAQMDD283 pKa = 3.68YY284 pKa = 7.44WTQMALTPLHH294 pKa = 5.34NTINHH299 pKa = 6.63FLRR302 pKa = 11.84ALKK305 pKa = 10.43EE306 pKa = 4.21DD307 pKa = 3.65GTFNQHH313 pKa = 7.04AIAEE317 pKa = 4.48RR318 pKa = 11.84VRR320 pKa = 11.84QWTADD325 pKa = 3.04PSMEE329 pKa = 4.14VFSFDD334 pKa = 3.07LTAATDD340 pKa = 3.63RR341 pKa = 11.84VPITFQEE348 pKa = 4.57SILSYY353 pKa = 11.4LMTSKK358 pKa = 10.86SFGNGWASILVDD370 pKa = 4.94RR371 pKa = 11.84EE372 pKa = 3.87FLTPNGDD379 pKa = 3.92LISYY383 pKa = 7.6NTGQPMGARR392 pKa = 11.84SSFPMLALTHH402 pKa = 6.48HH403 pKa = 7.23IIVQIAAARR412 pKa = 11.84AGLTVYY418 pKa = 10.44RR419 pKa = 11.84DD420 pKa = 3.73YY421 pKa = 11.74VVLGDD426 pKa = 6.02DD427 pKa = 3.61VTLTNAQVAAHH438 pKa = 6.11YY439 pKa = 7.21QTIMRR444 pKa = 11.84CLGVPINLSKK454 pKa = 11.05SIVHH458 pKa = 5.82VDD460 pKa = 3.32GGVSMAEE467 pKa = 3.23ICKK470 pKa = 9.85RR471 pKa = 11.84VFMDD475 pKa = 3.91GVEE478 pKa = 3.91ISRR481 pKa = 11.84FNPKK485 pKa = 10.33LIVNVIRR492 pKa = 11.84DD493 pKa = 3.74GRR495 pKa = 11.84LGPDD499 pKa = 3.7LQNDD503 pKa = 4.77LIIRR507 pKa = 11.84GWDD510 pKa = 3.29PSNEE514 pKa = 4.14VFWKK518 pKa = 10.25FMAGLLSIDD527 pKa = 4.01NLTLLIRR534 pKa = 11.84LNCAPISITGLLRR547 pKa = 11.84QFASNSKK554 pKa = 9.73LAQLSAWIPAYY565 pKa = 10.19QDD567 pKa = 3.69LKK569 pKa = 10.82PEE571 pKa = 4.19HH572 pKa = 6.55LVEE575 pKa = 4.29LFTYY579 pKa = 8.26VTASEE584 pKa = 3.96ALKK587 pKa = 10.76RR588 pKa = 11.84LDD590 pKa = 4.43GILRR594 pKa = 11.84AAVTINDD601 pKa = 3.94SLSIIAAANAHH612 pKa = 7.13PDD614 pKa = 4.05RR615 pKa = 11.84IPQYY619 pKa = 11.31VRR621 pKa = 11.84DD622 pKa = 3.61TWLGEE627 pKa = 3.86GLTKK631 pKa = 10.28EE632 pKa = 3.75EE633 pKa = 4.12RR634 pKa = 11.84ARR636 pKa = 11.84LEE638 pKa = 3.79GLIASTGPITPNHH651 pKa = 6.42PLVSASRR658 pKa = 11.84AEE660 pKa = 3.93ANRR663 pKa = 11.84ISEE666 pKa = 4.98LLHH669 pKa = 5.75QLNSHH674 pKa = 6.45DD675 pKa = 3.57TAIITRR681 pKa = 11.84ARR683 pKa = 11.84LGLLDD688 pKa = 3.58VFRR691 pKa = 11.84TSISSIWLDD700 pKa = 3.82DD701 pKa = 3.82GNIRR705 pKa = 11.84AGEE708 pKa = 4.02SRR710 pKa = 11.84SIFTRR715 pKa = 11.84MLTTLVSLFTSEE727 pKa = 4.23KK728 pKa = 9.76RR729 pKa = 11.84VSKK732 pKa = 10.52SGRR735 pKa = 11.84NLSLSYY741 pKa = 10.88SVVLTSLSRR750 pKa = 11.84LWTVALDD757 pKa = 3.61FGGQVTVNALRR768 pKa = 11.84ANVTRR773 pKa = 11.84DD774 pKa = 2.63IHH776 pKa = 6.35NAVDD780 pKa = 3.5NLKK783 pKa = 10.35AAEE786 pKa = 4.0EE787 pKa = 4.01AAVLISSSSVPTTSPTPATPKK808 pKa = 9.6GIRR811 pKa = 11.84RR812 pKa = 11.84RR813 pKa = 11.84RR814 pKa = 11.84AFARR818 pKa = 11.84ISS820 pKa = 3.22
Molecular weight: 91.19 kDa
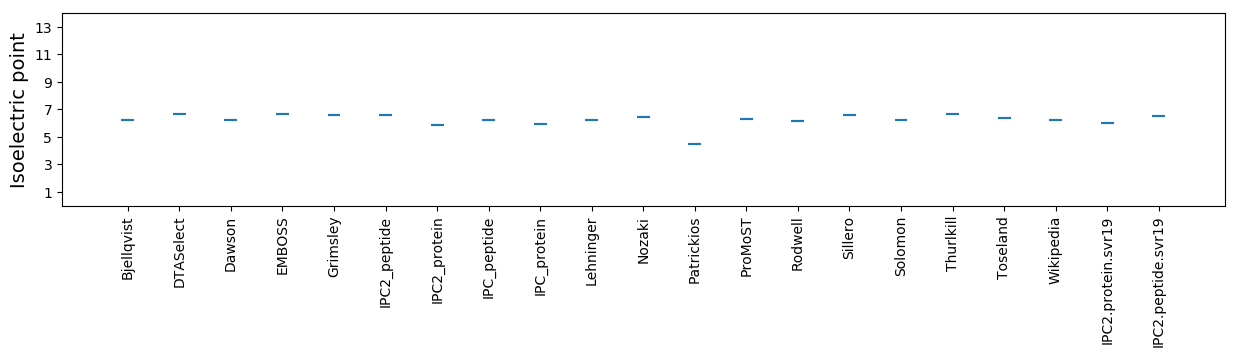
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E2RWR3|E2RWR3_9VIRU RNA-directed RNA polymerase OS=Rhizophagus sp. RF1 mitovirus OX=758866 GN=RdRp PE=4 SV=2
MM1 pKa = 7.15STFNFNSPNEE11 pKa = 4.04VRR13 pKa = 11.84RR14 pKa = 11.84VTRR17 pKa = 11.84SNIASLFAALMHH29 pKa = 6.55LNSLVTVTYY38 pKa = 9.13SQRR41 pKa = 11.84LLAFLGAVYY50 pKa = 10.82NRR52 pKa = 11.84ILVLFDD58 pKa = 3.76ANARR62 pKa = 11.84SLISEE67 pKa = 4.59LKK69 pKa = 10.32LVRR72 pKa = 11.84RR73 pKa = 11.84WFLEE77 pKa = 4.45FIRR80 pKa = 11.84NGNTDD85 pKa = 3.39NPGLEE90 pKa = 3.86WDD92 pKa = 4.1RR93 pKa = 11.84WDD95 pKa = 5.14DD96 pKa = 4.2SNNCPVLLEE105 pKa = 4.52GLDD108 pKa = 3.85ALWDD112 pKa = 4.55EE113 pKa = 4.94IEE115 pKa = 4.56SPSDD119 pKa = 3.12SHH121 pKa = 5.98YY122 pKa = 9.92TDD124 pKa = 4.25HH125 pKa = 7.42AAQLIFTLLSIDD137 pKa = 4.15RR138 pKa = 11.84IIVVPAVPNYY148 pKa = 9.48STIEE152 pKa = 4.35DD153 pKa = 4.19GPVIEE158 pKa = 4.36PVGYY162 pKa = 6.95PTNEE166 pKa = 3.85EE167 pKa = 4.02LVNALSSLHH176 pKa = 6.74IDD178 pKa = 3.4PAAFKK183 pKa = 10.73AFYY186 pKa = 9.08NQQVHH191 pKa = 6.66DD192 pKa = 4.22FDD194 pKa = 5.39YY195 pKa = 11.22EE196 pKa = 4.3VLSTRR201 pKa = 11.84GPNGDD206 pKa = 3.94ATWTAHH212 pKa = 6.22LDD214 pKa = 3.31ARR216 pKa = 11.84AWALDD221 pKa = 3.42TEE223 pKa = 4.5LFRR226 pKa = 11.84RR227 pKa = 11.84FSAWLEE233 pKa = 3.76EE234 pKa = 4.16SRR236 pKa = 11.84LTRR239 pKa = 11.84ILRR242 pKa = 11.84DD243 pKa = 3.17LFGCIRR249 pKa = 11.84SAAAEE254 pKa = 4.66AIPNLSPILGKK265 pKa = 10.73LSVIEE270 pKa = 4.01EE271 pKa = 4.27WGGKK275 pKa = 9.01ARR277 pKa = 11.84IVAQMDD283 pKa = 3.68YY284 pKa = 7.44WTQMALTPLHH294 pKa = 5.34NTINHH299 pKa = 6.63FLRR302 pKa = 11.84ALKK305 pKa = 10.43EE306 pKa = 4.21DD307 pKa = 3.65GTFNQHH313 pKa = 7.04AIAEE317 pKa = 4.48RR318 pKa = 11.84VRR320 pKa = 11.84QWTADD325 pKa = 3.04PSMEE329 pKa = 4.14VFSFDD334 pKa = 3.07LTAATDD340 pKa = 3.63RR341 pKa = 11.84VPITFQEE348 pKa = 4.57SILSYY353 pKa = 11.4LMTSKK358 pKa = 10.86SFGNGWASILVDD370 pKa = 4.94RR371 pKa = 11.84EE372 pKa = 3.87FLTPNGDD379 pKa = 3.92LISYY383 pKa = 7.6NTGQPMGARR392 pKa = 11.84SSFPMLALTHH402 pKa = 6.48HH403 pKa = 7.23IIVQIAAARR412 pKa = 11.84AGLTVYY418 pKa = 10.44RR419 pKa = 11.84DD420 pKa = 3.73YY421 pKa = 11.74VVLGDD426 pKa = 6.02DD427 pKa = 3.61VTLTNAQVAAHH438 pKa = 6.11YY439 pKa = 7.21QTIMRR444 pKa = 11.84CLGVPINLSKK454 pKa = 11.05SIVHH458 pKa = 5.82VDD460 pKa = 3.32GGVSMAEE467 pKa = 3.23ICKK470 pKa = 9.85RR471 pKa = 11.84VFMDD475 pKa = 3.91GVEE478 pKa = 3.91ISRR481 pKa = 11.84FNPKK485 pKa = 10.33LIVNVIRR492 pKa = 11.84DD493 pKa = 3.74GRR495 pKa = 11.84LGPDD499 pKa = 3.7LQNDD503 pKa = 4.77LIIRR507 pKa = 11.84GWDD510 pKa = 3.29PSNEE514 pKa = 4.14VFWKK518 pKa = 10.25FMAGLLSIDD527 pKa = 4.01NLTLLIRR534 pKa = 11.84LNCAPISITGLLRR547 pKa = 11.84QFASNSKK554 pKa = 9.73LAQLSAWIPAYY565 pKa = 10.19QDD567 pKa = 3.69LKK569 pKa = 10.82PEE571 pKa = 4.19HH572 pKa = 6.55LVEE575 pKa = 4.29LFTYY579 pKa = 8.26VTASEE584 pKa = 3.96ALKK587 pKa = 10.76RR588 pKa = 11.84LDD590 pKa = 4.43GILRR594 pKa = 11.84AAVTINDD601 pKa = 3.94SLSIIAAANAHH612 pKa = 7.13PDD614 pKa = 4.05RR615 pKa = 11.84IPQYY619 pKa = 11.31VRR621 pKa = 11.84DD622 pKa = 3.61TWLGEE627 pKa = 3.86GLTKK631 pKa = 10.28EE632 pKa = 3.75EE633 pKa = 4.12RR634 pKa = 11.84ARR636 pKa = 11.84LEE638 pKa = 3.79GLIASTGPITPNHH651 pKa = 6.42PLVSASRR658 pKa = 11.84AEE660 pKa = 3.93ANRR663 pKa = 11.84ISEE666 pKa = 4.98LLHH669 pKa = 5.75QLNSHH674 pKa = 6.45DD675 pKa = 3.57TAIITRR681 pKa = 11.84ARR683 pKa = 11.84LGLLDD688 pKa = 3.58VFRR691 pKa = 11.84TSISSIWLDD700 pKa = 3.82DD701 pKa = 3.82GNIRR705 pKa = 11.84AGEE708 pKa = 4.02SRR710 pKa = 11.84SIFTRR715 pKa = 11.84MLTTLVSLFTSEE727 pKa = 4.23KK728 pKa = 9.76RR729 pKa = 11.84VSKK732 pKa = 10.52SGRR735 pKa = 11.84NLSLSYY741 pKa = 10.88SVVLTSLSRR750 pKa = 11.84LWTVALDD757 pKa = 3.61FGGQVTVNALRR768 pKa = 11.84ANVTRR773 pKa = 11.84DD774 pKa = 2.63IHH776 pKa = 6.35NAVDD780 pKa = 3.5NLKK783 pKa = 10.35AAEE786 pKa = 4.0EE787 pKa = 4.01AAVLISSSSVPTTSPTPATPKK808 pKa = 9.6GIRR811 pKa = 11.84RR812 pKa = 11.84RR813 pKa = 11.84RR814 pKa = 11.84AFARR818 pKa = 11.84ISS820 pKa = 3.22
MM1 pKa = 7.15STFNFNSPNEE11 pKa = 4.04VRR13 pKa = 11.84RR14 pKa = 11.84VTRR17 pKa = 11.84SNIASLFAALMHH29 pKa = 6.55LNSLVTVTYY38 pKa = 9.13SQRR41 pKa = 11.84LLAFLGAVYY50 pKa = 10.82NRR52 pKa = 11.84ILVLFDD58 pKa = 3.76ANARR62 pKa = 11.84SLISEE67 pKa = 4.59LKK69 pKa = 10.32LVRR72 pKa = 11.84RR73 pKa = 11.84WFLEE77 pKa = 4.45FIRR80 pKa = 11.84NGNTDD85 pKa = 3.39NPGLEE90 pKa = 3.86WDD92 pKa = 4.1RR93 pKa = 11.84WDD95 pKa = 5.14DD96 pKa = 4.2SNNCPVLLEE105 pKa = 4.52GLDD108 pKa = 3.85ALWDD112 pKa = 4.55EE113 pKa = 4.94IEE115 pKa = 4.56SPSDD119 pKa = 3.12SHH121 pKa = 5.98YY122 pKa = 9.92TDD124 pKa = 4.25HH125 pKa = 7.42AAQLIFTLLSIDD137 pKa = 4.15RR138 pKa = 11.84IIVVPAVPNYY148 pKa = 9.48STIEE152 pKa = 4.35DD153 pKa = 4.19GPVIEE158 pKa = 4.36PVGYY162 pKa = 6.95PTNEE166 pKa = 3.85EE167 pKa = 4.02LVNALSSLHH176 pKa = 6.74IDD178 pKa = 3.4PAAFKK183 pKa = 10.73AFYY186 pKa = 9.08NQQVHH191 pKa = 6.66DD192 pKa = 4.22FDD194 pKa = 5.39YY195 pKa = 11.22EE196 pKa = 4.3VLSTRR201 pKa = 11.84GPNGDD206 pKa = 3.94ATWTAHH212 pKa = 6.22LDD214 pKa = 3.31ARR216 pKa = 11.84AWALDD221 pKa = 3.42TEE223 pKa = 4.5LFRR226 pKa = 11.84RR227 pKa = 11.84FSAWLEE233 pKa = 3.76EE234 pKa = 4.16SRR236 pKa = 11.84LTRR239 pKa = 11.84ILRR242 pKa = 11.84DD243 pKa = 3.17LFGCIRR249 pKa = 11.84SAAAEE254 pKa = 4.66AIPNLSPILGKK265 pKa = 10.73LSVIEE270 pKa = 4.01EE271 pKa = 4.27WGGKK275 pKa = 9.01ARR277 pKa = 11.84IVAQMDD283 pKa = 3.68YY284 pKa = 7.44WTQMALTPLHH294 pKa = 5.34NTINHH299 pKa = 6.63FLRR302 pKa = 11.84ALKK305 pKa = 10.43EE306 pKa = 4.21DD307 pKa = 3.65GTFNQHH313 pKa = 7.04AIAEE317 pKa = 4.48RR318 pKa = 11.84VRR320 pKa = 11.84QWTADD325 pKa = 3.04PSMEE329 pKa = 4.14VFSFDD334 pKa = 3.07LTAATDD340 pKa = 3.63RR341 pKa = 11.84VPITFQEE348 pKa = 4.57SILSYY353 pKa = 11.4LMTSKK358 pKa = 10.86SFGNGWASILVDD370 pKa = 4.94RR371 pKa = 11.84EE372 pKa = 3.87FLTPNGDD379 pKa = 3.92LISYY383 pKa = 7.6NTGQPMGARR392 pKa = 11.84SSFPMLALTHH402 pKa = 6.48HH403 pKa = 7.23IIVQIAAARR412 pKa = 11.84AGLTVYY418 pKa = 10.44RR419 pKa = 11.84DD420 pKa = 3.73YY421 pKa = 11.74VVLGDD426 pKa = 6.02DD427 pKa = 3.61VTLTNAQVAAHH438 pKa = 6.11YY439 pKa = 7.21QTIMRR444 pKa = 11.84CLGVPINLSKK454 pKa = 11.05SIVHH458 pKa = 5.82VDD460 pKa = 3.32GGVSMAEE467 pKa = 3.23ICKK470 pKa = 9.85RR471 pKa = 11.84VFMDD475 pKa = 3.91GVEE478 pKa = 3.91ISRR481 pKa = 11.84FNPKK485 pKa = 10.33LIVNVIRR492 pKa = 11.84DD493 pKa = 3.74GRR495 pKa = 11.84LGPDD499 pKa = 3.7LQNDD503 pKa = 4.77LIIRR507 pKa = 11.84GWDD510 pKa = 3.29PSNEE514 pKa = 4.14VFWKK518 pKa = 10.25FMAGLLSIDD527 pKa = 4.01NLTLLIRR534 pKa = 11.84LNCAPISITGLLRR547 pKa = 11.84QFASNSKK554 pKa = 9.73LAQLSAWIPAYY565 pKa = 10.19QDD567 pKa = 3.69LKK569 pKa = 10.82PEE571 pKa = 4.19HH572 pKa = 6.55LVEE575 pKa = 4.29LFTYY579 pKa = 8.26VTASEE584 pKa = 3.96ALKK587 pKa = 10.76RR588 pKa = 11.84LDD590 pKa = 4.43GILRR594 pKa = 11.84AAVTINDD601 pKa = 3.94SLSIIAAANAHH612 pKa = 7.13PDD614 pKa = 4.05RR615 pKa = 11.84IPQYY619 pKa = 11.31VRR621 pKa = 11.84DD622 pKa = 3.61TWLGEE627 pKa = 3.86GLTKK631 pKa = 10.28EE632 pKa = 3.75EE633 pKa = 4.12RR634 pKa = 11.84ARR636 pKa = 11.84LEE638 pKa = 3.79GLIASTGPITPNHH651 pKa = 6.42PLVSASRR658 pKa = 11.84AEE660 pKa = 3.93ANRR663 pKa = 11.84ISEE666 pKa = 4.98LLHH669 pKa = 5.75QLNSHH674 pKa = 6.45DD675 pKa = 3.57TAIITRR681 pKa = 11.84ARR683 pKa = 11.84LGLLDD688 pKa = 3.58VFRR691 pKa = 11.84TSISSIWLDD700 pKa = 3.82DD701 pKa = 3.82GNIRR705 pKa = 11.84AGEE708 pKa = 4.02SRR710 pKa = 11.84SIFTRR715 pKa = 11.84MLTTLVSLFTSEE727 pKa = 4.23KK728 pKa = 9.76RR729 pKa = 11.84VSKK732 pKa = 10.52SGRR735 pKa = 11.84NLSLSYY741 pKa = 10.88SVVLTSLSRR750 pKa = 11.84LWTVALDD757 pKa = 3.61FGGQVTVNALRR768 pKa = 11.84ANVTRR773 pKa = 11.84DD774 pKa = 2.63IHH776 pKa = 6.35NAVDD780 pKa = 3.5NLKK783 pKa = 10.35AAEE786 pKa = 4.0EE787 pKa = 4.01AAVLISSSSVPTTSPTPATPKK808 pKa = 9.6GIRR811 pKa = 11.84RR812 pKa = 11.84RR813 pKa = 11.84RR814 pKa = 11.84AFARR818 pKa = 11.84ISS820 pKa = 3.22
Molecular weight: 91.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
820 |
820 |
820 |
820.0 |
91.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.512 ± 0.0 | 0.61 ± 0.0 |
5.732 ± 0.0 | 4.756 ± 0.0 |
4.024 ± 0.0 | 5.0 ± 0.0 |
2.317 ± 0.0 | 7.195 ± 0.0 |
2.195 ± 0.0 | 11.829 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.585 ± 0.0 | 5.366 ± 0.0 |
4.268 ± 0.0 | 2.439 ± 0.0 |
7.317 ± 0.0 | 8.537 ± 0.0 |
6.585 ± 0.0 | 6.585 ± 0.0 |
2.073 ± 0.0 | 2.073 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
