
Actinoplanes friuliensis DSM 7358
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micromonosporales; Micromonosporaceae; Actinoplanes; Actinoplanes friuliensis
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
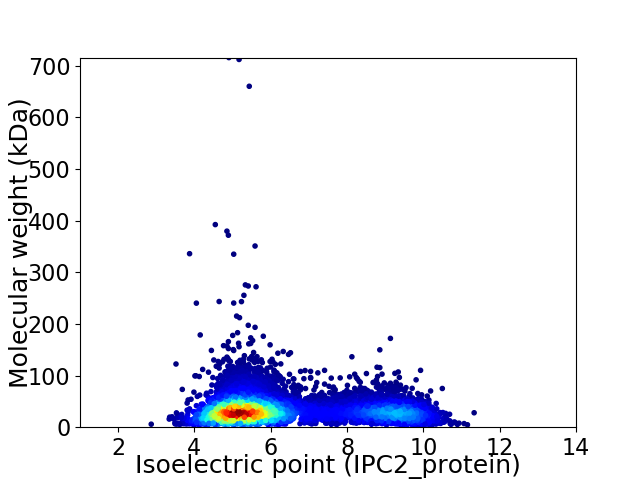
Virtual 2D-PAGE plot for 8558 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
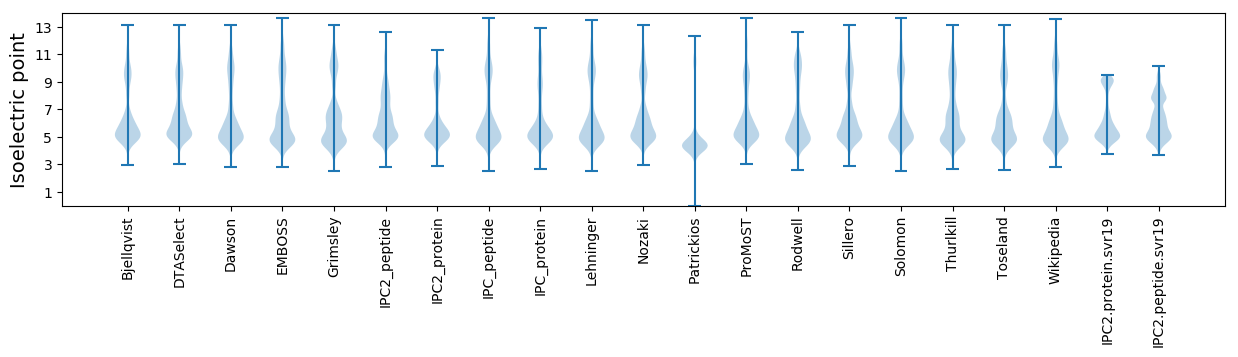
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U5VU47|U5VU47_9ACTN Putative MFS transporter OS=Actinoplanes friuliensis DSM 7358 OX=1246995 GN=AFR_04285 PE=4 SV=1
MM1 pKa = 8.25DD2 pKa = 6.16DD3 pKa = 3.3PTAFLTEE10 pKa = 3.98PAKK13 pKa = 10.78PDD15 pKa = 3.58DD16 pKa = 3.99SVSNGFANPLDD27 pKa = 3.7LFNYY31 pKa = 8.8VSPSAWLNAAIEE43 pKa = 4.32KK44 pKa = 9.13LTGVDD49 pKa = 3.24VFGWMTDD56 pKa = 3.01WVSGDD61 pKa = 3.39WEE63 pKa = 4.61SLWKK67 pKa = 11.14YY68 pKa = 9.82GDD70 pKa = 3.32AMANLAQCMQQIGINIQTGMLQLDD94 pKa = 4.05ASWDD98 pKa = 3.87GNASDD103 pKa = 4.66AAYY106 pKa = 9.66KK107 pKa = 10.65YY108 pKa = 10.94FSDD111 pKa = 4.55LAAATSGQQVAIAKK125 pKa = 7.75TQDD128 pKa = 3.56SYY130 pKa = 12.12HH131 pKa = 6.45KK132 pKa = 9.98AALGAWQLSNQLGNILQALADD153 pKa = 3.87KK154 pKa = 10.92AILAGIAAAAGTALIEE170 pKa = 4.34TGVGAVAGYY179 pKa = 9.43GAAAYY184 pKa = 9.25IVLDD188 pKa = 3.8MLQLINDD195 pKa = 3.94ASVIINTAGTVILGLFGGVMDD216 pKa = 5.47AAYY219 pKa = 10.42QGGDD223 pKa = 3.42LTAVPLPATAYY234 pKa = 9.99SGPGAA239 pKa = 4.07
MM1 pKa = 8.25DD2 pKa = 6.16DD3 pKa = 3.3PTAFLTEE10 pKa = 3.98PAKK13 pKa = 10.78PDD15 pKa = 3.58DD16 pKa = 3.99SVSNGFANPLDD27 pKa = 3.7LFNYY31 pKa = 8.8VSPSAWLNAAIEE43 pKa = 4.32KK44 pKa = 9.13LTGVDD49 pKa = 3.24VFGWMTDD56 pKa = 3.01WVSGDD61 pKa = 3.39WEE63 pKa = 4.61SLWKK67 pKa = 11.14YY68 pKa = 9.82GDD70 pKa = 3.32AMANLAQCMQQIGINIQTGMLQLDD94 pKa = 4.05ASWDD98 pKa = 3.87GNASDD103 pKa = 4.66AAYY106 pKa = 9.66KK107 pKa = 10.65YY108 pKa = 10.94FSDD111 pKa = 4.55LAAATSGQQVAIAKK125 pKa = 7.75TQDD128 pKa = 3.56SYY130 pKa = 12.12HH131 pKa = 6.45KK132 pKa = 9.98AALGAWQLSNQLGNILQALADD153 pKa = 3.87KK154 pKa = 10.92AILAGIAAAAGTALIEE170 pKa = 4.34TGVGAVAGYY179 pKa = 9.43GAAAYY184 pKa = 9.25IVLDD188 pKa = 3.8MLQLINDD195 pKa = 3.94ASVIINTAGTVILGLFGGVMDD216 pKa = 5.47AAYY219 pKa = 10.42QGGDD223 pKa = 3.42LTAVPLPATAYY234 pKa = 9.99SGPGAA239 pKa = 4.07
Molecular weight: 24.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U5W634|U5W634_9ACTN YceI domain-containing protein OS=Actinoplanes friuliensis DSM 7358 OX=1246995 GN=AFR_31890 PE=4 SV=1
MM1 pKa = 7.07TRR3 pKa = 11.84RR4 pKa = 11.84GGISRR9 pKa = 11.84IRR11 pKa = 11.84NGHH14 pKa = 5.22TGGPATGRR22 pKa = 11.84PDD24 pKa = 3.45PRR26 pKa = 11.84LTAIDD31 pKa = 3.55RR32 pKa = 11.84RR33 pKa = 11.84MPASGLLRR41 pKa = 11.84DD42 pKa = 3.76TTPPRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84LTRR52 pKa = 11.84RR53 pKa = 11.84SLSGHH58 pKa = 5.81GLTRR62 pKa = 11.84HH63 pKa = 5.37RR64 pKa = 11.84TTSRR68 pKa = 11.84RR69 pKa = 11.84LTRR72 pKa = 11.84HH73 pKa = 5.27GLTWHH78 pKa = 6.18WLARR82 pKa = 11.84HH83 pKa = 6.21RR84 pKa = 11.84LPGNGLARR92 pKa = 11.84SRR94 pKa = 11.84TTRR97 pKa = 11.84HH98 pKa = 5.64GLTLTGTAGRR108 pKa = 11.84GLTRR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84VTGPGLTRR122 pKa = 11.84TGTTRR127 pKa = 11.84HH128 pKa = 5.82SLTSRR133 pKa = 11.84SMTSPRR139 pKa = 11.84LTRR142 pKa = 11.84TWTTRR147 pKa = 11.84HH148 pKa = 5.84GLTSSSMTGGGLTRR162 pKa = 11.84TRR164 pKa = 11.84TTRR167 pKa = 11.84HH168 pKa = 5.48GLTSRR173 pKa = 11.84SMTGRR178 pKa = 11.84GLTRR182 pKa = 11.84NGTTRR187 pKa = 11.84NGLTSRR193 pKa = 11.84SLTRR197 pKa = 11.84PRR199 pKa = 11.84LTRR202 pKa = 11.84TWTTRR207 pKa = 11.84HH208 pKa = 5.38GLTRR212 pKa = 11.84SGLTLNRR219 pKa = 11.84TGTAGRR225 pKa = 11.84GLAGRR230 pKa = 11.84RR231 pKa = 11.84VTGRR235 pKa = 11.84GLTRR239 pKa = 11.84ARR241 pKa = 11.84TTRR244 pKa = 11.84HH245 pKa = 5.49SLTSRR250 pKa = 11.84SMTT253 pKa = 3.47
MM1 pKa = 7.07TRR3 pKa = 11.84RR4 pKa = 11.84GGISRR9 pKa = 11.84IRR11 pKa = 11.84NGHH14 pKa = 5.22TGGPATGRR22 pKa = 11.84PDD24 pKa = 3.45PRR26 pKa = 11.84LTAIDD31 pKa = 3.55RR32 pKa = 11.84RR33 pKa = 11.84MPASGLLRR41 pKa = 11.84DD42 pKa = 3.76TTPPRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84LTRR52 pKa = 11.84RR53 pKa = 11.84SLSGHH58 pKa = 5.81GLTRR62 pKa = 11.84HH63 pKa = 5.37RR64 pKa = 11.84TTSRR68 pKa = 11.84RR69 pKa = 11.84LTRR72 pKa = 11.84HH73 pKa = 5.27GLTWHH78 pKa = 6.18WLARR82 pKa = 11.84HH83 pKa = 6.21RR84 pKa = 11.84LPGNGLARR92 pKa = 11.84SRR94 pKa = 11.84TTRR97 pKa = 11.84HH98 pKa = 5.64GLTLTGTAGRR108 pKa = 11.84GLTRR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84VTGPGLTRR122 pKa = 11.84TGTTRR127 pKa = 11.84HH128 pKa = 5.82SLTSRR133 pKa = 11.84SMTSPRR139 pKa = 11.84LTRR142 pKa = 11.84TWTTRR147 pKa = 11.84HH148 pKa = 5.84GLTSSSMTGGGLTRR162 pKa = 11.84TRR164 pKa = 11.84TTRR167 pKa = 11.84HH168 pKa = 5.48GLTSRR173 pKa = 11.84SMTGRR178 pKa = 11.84GLTRR182 pKa = 11.84NGTTRR187 pKa = 11.84NGLTSRR193 pKa = 11.84SLTRR197 pKa = 11.84PRR199 pKa = 11.84LTRR202 pKa = 11.84TWTTRR207 pKa = 11.84HH208 pKa = 5.38GLTRR212 pKa = 11.84SGLTLNRR219 pKa = 11.84TGTAGRR225 pKa = 11.84GLAGRR230 pKa = 11.84RR231 pKa = 11.84VTGRR235 pKa = 11.84GLTRR239 pKa = 11.84ARR241 pKa = 11.84TTRR244 pKa = 11.84HH245 pKa = 5.49SLTSRR250 pKa = 11.84SMTT253 pKa = 3.47
Molecular weight: 27.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2802956 |
29 |
6661 |
327.5 |
34.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.646 ± 0.038 | 0.659 ± 0.007 |
6.074 ± 0.023 | 5.094 ± 0.026 |
2.836 ± 0.015 | 9.262 ± 0.029 |
2.028 ± 0.012 | 3.608 ± 0.019 |
2.004 ± 0.022 | 10.532 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.685 ± 0.009 | 1.981 ± 0.019 |
5.983 ± 0.024 | 2.847 ± 0.018 |
7.659 ± 0.031 | 5.194 ± 0.023 |
6.488 ± 0.032 | 8.784 ± 0.025 |
1.568 ± 0.012 | 2.067 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
