
CRESS virus sp. ct6pe1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; unclassified Cressdnaviricota; CRESS viruses
Average proteome isoelectric point is 8.16
Get precalculated fractions of proteins
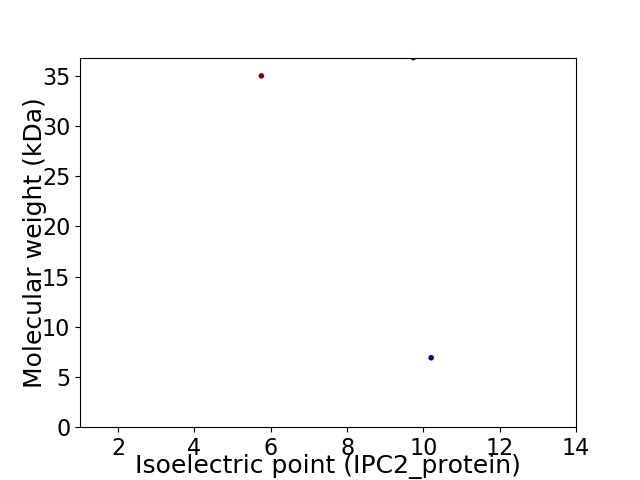
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2WBB5|A0A5Q2WBB5_9VIRU Rep protein OS=CRESS virus sp. ct6pe1 OX=2656676 PE=4 SV=1
MM1 pKa = 7.7PAAQARR7 pKa = 11.84RR8 pKa = 11.84FCFTLNNYY16 pKa = 7.93TPEE19 pKa = 4.29DD20 pKa = 3.67VAHH23 pKa = 6.9IEE25 pKa = 4.17SLYY28 pKa = 11.06LNADD32 pKa = 3.01NHH34 pKa = 5.06ITYY37 pKa = 10.37LVIGRR42 pKa = 11.84EE43 pKa = 4.33VGDD46 pKa = 3.43SGTPHH51 pKa = 6.38LQGFICFSRR60 pKa = 11.84RR61 pKa = 11.84KK62 pKa = 7.52TFNVVKK68 pKa = 10.57ALINEE73 pKa = 4.98RR74 pKa = 11.84IHH76 pKa = 8.47IEE78 pKa = 4.06GARR81 pKa = 11.84GTSDD85 pKa = 3.86QAATYY90 pKa = 9.19CKK92 pKa = 10.12KK93 pKa = 10.65DD94 pKa = 3.08GAYY97 pKa = 9.27TEE99 pKa = 4.8FGTVPVSGKK108 pKa = 8.36TNYY111 pKa = 10.7LEE113 pKa = 6.47DD114 pKa = 3.86FFKK117 pKa = 10.56WADD120 pKa = 3.48QFHH123 pKa = 7.33EE124 pKa = 4.73DD125 pKa = 3.63NGRR128 pKa = 11.84IPTARR133 pKa = 11.84DD134 pKa = 2.93VALEE138 pKa = 4.04HH139 pKa = 6.43PVVLTRR145 pKa = 11.84HH146 pKa = 6.03RR147 pKa = 11.84NVLDD151 pKa = 3.54VLEE154 pKa = 4.87SRR156 pKa = 11.84APPPSLVTNPQPRR169 pKa = 11.84PWQLEE174 pKa = 4.29LEE176 pKa = 4.28EE177 pKa = 5.29HH178 pKa = 6.16LTDD181 pKa = 4.54DD182 pKa = 4.05TPDD185 pKa = 3.4DD186 pKa = 3.59RR187 pKa = 11.84TVEE190 pKa = 4.05FFVDD194 pKa = 3.56EE195 pKa = 4.36EE196 pKa = 4.52GGKK199 pKa = 10.11GKK201 pKa = 10.14SWFQRR206 pKa = 11.84HH207 pKa = 5.99MLTKK211 pKa = 10.5YY212 pKa = 9.53PDD214 pKa = 3.87RR215 pKa = 11.84VQMLAPGKK223 pKa = 10.08RR224 pKa = 11.84DD225 pKa = 3.9DD226 pKa = 4.31LAHH229 pKa = 6.75TIDD232 pKa = 3.36IQKK235 pKa = 10.71SIFLMNVPKK244 pKa = 9.55TQMEE248 pKa = 4.36FLQYY252 pKa = 10.87SILEE256 pKa = 4.06QLKK259 pKa = 10.27DD260 pKa = 3.4RR261 pKa = 11.84TVFSPKK267 pKa = 8.73YY268 pKa = 7.05TSKK271 pKa = 10.38MKK273 pKa = 10.61VLSAPAHH280 pKa = 5.4VIVFSNEE287 pKa = 3.72HH288 pKa = 6.41PDD290 pKa = 3.72MNKK293 pKa = 9.18MSADD297 pKa = 3.78RR298 pKa = 11.84YY299 pKa = 10.49HH300 pKa = 6.34ITILDD305 pKa = 3.51
MM1 pKa = 7.7PAAQARR7 pKa = 11.84RR8 pKa = 11.84FCFTLNNYY16 pKa = 7.93TPEE19 pKa = 4.29DD20 pKa = 3.67VAHH23 pKa = 6.9IEE25 pKa = 4.17SLYY28 pKa = 11.06LNADD32 pKa = 3.01NHH34 pKa = 5.06ITYY37 pKa = 10.37LVIGRR42 pKa = 11.84EE43 pKa = 4.33VGDD46 pKa = 3.43SGTPHH51 pKa = 6.38LQGFICFSRR60 pKa = 11.84RR61 pKa = 11.84KK62 pKa = 7.52TFNVVKK68 pKa = 10.57ALINEE73 pKa = 4.98RR74 pKa = 11.84IHH76 pKa = 8.47IEE78 pKa = 4.06GARR81 pKa = 11.84GTSDD85 pKa = 3.86QAATYY90 pKa = 9.19CKK92 pKa = 10.12KK93 pKa = 10.65DD94 pKa = 3.08GAYY97 pKa = 9.27TEE99 pKa = 4.8FGTVPVSGKK108 pKa = 8.36TNYY111 pKa = 10.7LEE113 pKa = 6.47DD114 pKa = 3.86FFKK117 pKa = 10.56WADD120 pKa = 3.48QFHH123 pKa = 7.33EE124 pKa = 4.73DD125 pKa = 3.63NGRR128 pKa = 11.84IPTARR133 pKa = 11.84DD134 pKa = 2.93VALEE138 pKa = 4.04HH139 pKa = 6.43PVVLTRR145 pKa = 11.84HH146 pKa = 6.03RR147 pKa = 11.84NVLDD151 pKa = 3.54VLEE154 pKa = 4.87SRR156 pKa = 11.84APPPSLVTNPQPRR169 pKa = 11.84PWQLEE174 pKa = 4.29LEE176 pKa = 4.28EE177 pKa = 5.29HH178 pKa = 6.16LTDD181 pKa = 4.54DD182 pKa = 4.05TPDD185 pKa = 3.4DD186 pKa = 3.59RR187 pKa = 11.84TVEE190 pKa = 4.05FFVDD194 pKa = 3.56EE195 pKa = 4.36EE196 pKa = 4.52GGKK199 pKa = 10.11GKK201 pKa = 10.14SWFQRR206 pKa = 11.84HH207 pKa = 5.99MLTKK211 pKa = 10.5YY212 pKa = 9.53PDD214 pKa = 3.87RR215 pKa = 11.84VQMLAPGKK223 pKa = 10.08RR224 pKa = 11.84DD225 pKa = 3.9DD226 pKa = 4.31LAHH229 pKa = 6.75TIDD232 pKa = 3.36IQKK235 pKa = 10.71SIFLMNVPKK244 pKa = 9.55TQMEE248 pKa = 4.36FLQYY252 pKa = 10.87SILEE256 pKa = 4.06QLKK259 pKa = 10.27DD260 pKa = 3.4RR261 pKa = 11.84TVFSPKK267 pKa = 8.73YY268 pKa = 7.05TSKK271 pKa = 10.38MKK273 pKa = 10.61VLSAPAHH280 pKa = 5.4VIVFSNEE287 pKa = 3.72HH288 pKa = 6.41PDD290 pKa = 3.72MNKK293 pKa = 9.18MSADD297 pKa = 3.78RR298 pKa = 11.84YY299 pKa = 10.49HH300 pKa = 6.34ITILDD305 pKa = 3.51
Molecular weight: 34.99 kDa
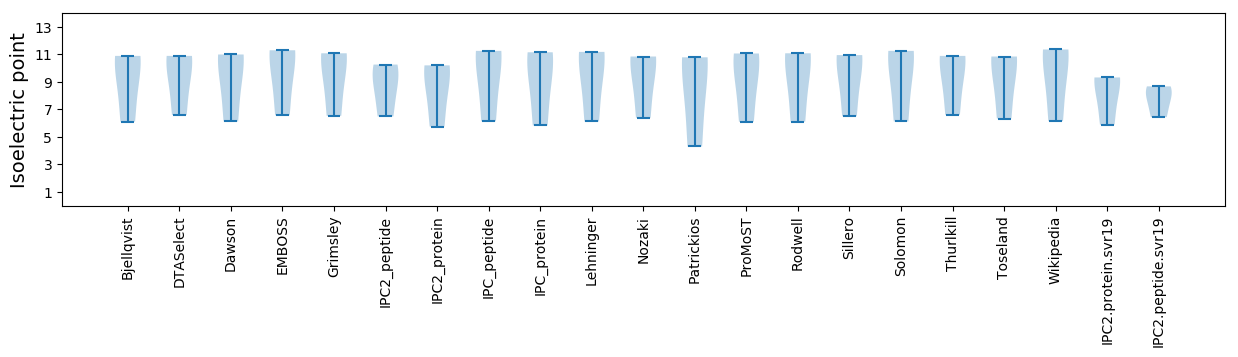
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2WBB5|A0A5Q2WBB5_9VIRU Rep protein OS=CRESS virus sp. ct6pe1 OX=2656676 PE=4 SV=1
MM1 pKa = 7.0KK2 pKa = 10.38HH3 pKa = 5.13NWTRR7 pKa = 11.84PIVKK11 pKa = 10.0FIEE14 pKa = 5.0DD15 pKa = 4.05DD16 pKa = 3.9LVASGIGGATGPARR30 pKa = 11.84FFAARR35 pKa = 11.84GAGLVGDD42 pKa = 4.99GVNKK46 pKa = 9.18TWGSGDD52 pKa = 3.27TRR54 pKa = 11.84TWSAWDD60 pKa = 3.26NMFVQKK66 pKa = 10.44SGQYY70 pKa = 7.36GQKK73 pKa = 9.64RR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84FTPTRR81 pKa = 11.84GKK83 pKa = 10.41RR84 pKa = 11.84GVRR87 pKa = 11.84LYY89 pKa = 9.71RR90 pKa = 11.84RR91 pKa = 11.84KK92 pKa = 10.18YY93 pKa = 7.88IRR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84PRR99 pKa = 11.84MMRR102 pKa = 11.84RR103 pKa = 11.84SRR105 pKa = 11.84ASWGTRR111 pKa = 11.84VRR113 pKa = 11.84RR114 pKa = 11.84AALRR118 pKa = 11.84SNEE121 pKa = 4.35SKK123 pKa = 10.22RR124 pKa = 11.84HH125 pKa = 4.09QVTTITEE132 pKa = 4.73AIDD135 pKa = 4.05DD136 pKa = 4.07QVLDD140 pKa = 3.75TTAIARR146 pKa = 11.84VPDD149 pKa = 4.18LDD151 pKa = 3.57QGAGGIEE158 pKa = 4.17GMSSKK163 pKa = 10.3ISRR166 pKa = 11.84NGATIITTGIASWIHH181 pKa = 5.52IRR183 pKa = 11.84NSSTVQAMRR192 pKa = 11.84VDD194 pKa = 4.28LVWFYY199 pKa = 11.66KK200 pKa = 10.49KK201 pKa = 10.14RR202 pKa = 11.84DD203 pKa = 3.58VGAVQAIFKK212 pKa = 10.21DD213 pKa = 3.8NQFEE217 pKa = 4.88AINSPSDD224 pKa = 3.31PTSYY228 pKa = 9.44FARR231 pKa = 11.84MIMPYY236 pKa = 10.88GNLNTVILKK245 pKa = 10.58KK246 pKa = 10.1LTIDD250 pKa = 3.36LAEE253 pKa = 4.71AGEE256 pKa = 4.42TAGTEE261 pKa = 3.96SRR263 pKa = 11.84MLKK266 pKa = 9.65VWLPFKK272 pKa = 10.8KK273 pKa = 10.45VMKK276 pKa = 10.49FNYY279 pKa = 8.95TSEE282 pKa = 3.99GTTNQDD288 pKa = 2.52YY289 pKa = 10.73DD290 pKa = 3.26IHH292 pKa = 8.03FGIRR296 pKa = 11.84CHH298 pKa = 6.14NKK300 pKa = 8.99EE301 pKa = 3.79GQAFSTTTNVLIAQHH316 pKa = 5.85RR317 pKa = 11.84HH318 pKa = 3.41VLYY321 pKa = 10.79FRR323 pKa = 11.84DD324 pKa = 3.69PP325 pKa = 3.42
MM1 pKa = 7.0KK2 pKa = 10.38HH3 pKa = 5.13NWTRR7 pKa = 11.84PIVKK11 pKa = 10.0FIEE14 pKa = 5.0DD15 pKa = 4.05DD16 pKa = 3.9LVASGIGGATGPARR30 pKa = 11.84FFAARR35 pKa = 11.84GAGLVGDD42 pKa = 4.99GVNKK46 pKa = 9.18TWGSGDD52 pKa = 3.27TRR54 pKa = 11.84TWSAWDD60 pKa = 3.26NMFVQKK66 pKa = 10.44SGQYY70 pKa = 7.36GQKK73 pKa = 9.64RR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84FTPTRR81 pKa = 11.84GKK83 pKa = 10.41RR84 pKa = 11.84GVRR87 pKa = 11.84LYY89 pKa = 9.71RR90 pKa = 11.84RR91 pKa = 11.84KK92 pKa = 10.18YY93 pKa = 7.88IRR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84PRR99 pKa = 11.84MMRR102 pKa = 11.84RR103 pKa = 11.84SRR105 pKa = 11.84ASWGTRR111 pKa = 11.84VRR113 pKa = 11.84RR114 pKa = 11.84AALRR118 pKa = 11.84SNEE121 pKa = 4.35SKK123 pKa = 10.22RR124 pKa = 11.84HH125 pKa = 4.09QVTTITEE132 pKa = 4.73AIDD135 pKa = 4.05DD136 pKa = 4.07QVLDD140 pKa = 3.75TTAIARR146 pKa = 11.84VPDD149 pKa = 4.18LDD151 pKa = 3.57QGAGGIEE158 pKa = 4.17GMSSKK163 pKa = 10.3ISRR166 pKa = 11.84NGATIITTGIASWIHH181 pKa = 5.52IRR183 pKa = 11.84NSSTVQAMRR192 pKa = 11.84VDD194 pKa = 4.28LVWFYY199 pKa = 11.66KK200 pKa = 10.49KK201 pKa = 10.14RR202 pKa = 11.84DD203 pKa = 3.58VGAVQAIFKK212 pKa = 10.21DD213 pKa = 3.8NQFEE217 pKa = 4.88AINSPSDD224 pKa = 3.31PTSYY228 pKa = 9.44FARR231 pKa = 11.84MIMPYY236 pKa = 10.88GNLNTVILKK245 pKa = 10.58KK246 pKa = 10.1LTIDD250 pKa = 3.36LAEE253 pKa = 4.71AGEE256 pKa = 4.42TAGTEE261 pKa = 3.96SRR263 pKa = 11.84MLKK266 pKa = 9.65VWLPFKK272 pKa = 10.8KK273 pKa = 10.45VMKK276 pKa = 10.49FNYY279 pKa = 8.95TSEE282 pKa = 3.99GTTNQDD288 pKa = 2.52YY289 pKa = 10.73DD290 pKa = 3.26IHH292 pKa = 8.03FGIRR296 pKa = 11.84CHH298 pKa = 6.14NKK300 pKa = 8.99EE301 pKa = 3.79GQAFSTTTNVLIAQHH316 pKa = 5.85RR317 pKa = 11.84HH318 pKa = 3.41VLYY321 pKa = 10.79FRR323 pKa = 11.84DD324 pKa = 3.69PP325 pKa = 3.42
Molecular weight: 36.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
691 |
61 |
325 |
230.3 |
26.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.946 ± 0.656 | 0.724 ± 0.263 |
6.078 ± 0.874 | 4.342 ± 1.029 |
4.631 ± 0.341 | 6.512 ± 1.3 |
3.039 ± 0.672 | 5.933 ± 0.576 |
5.355 ± 0.576 | 6.223 ± 1.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.039 ± 0.306 | 4.197 ± 0.141 |
4.631 ± 0.988 | 3.473 ± 0.524 |
8.828 ± 1.416 | 6.368 ± 1.291 |
8.104 ± 0.489 | 6.657 ± 0.341 |
1.592 ± 0.539 | 3.329 ± 0.527 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
