
Circoviridae sp. ctfvP2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.87
Get precalculated fractions of proteins
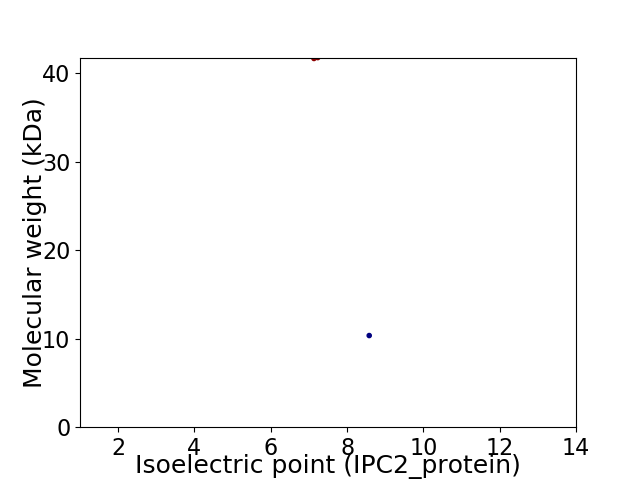
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2W9F3|A0A5Q2W9F3_9CIRC ATP-dependent helicase Rep OS=Circoviridae sp. ctfvP2 OX=2656728 PE=3 SV=1
MM1 pKa = 7.44EE2 pKa = 5.1GKK4 pKa = 9.88SIKK7 pKa = 9.6RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 10.14LFTLNKK17 pKa = 9.56PLEE20 pKa = 4.53KK21 pKa = 10.04KK22 pKa = 10.7DD23 pKa = 3.19IDD25 pKa = 3.94FNNNVVRR32 pKa = 11.84CYY34 pKa = 10.34CYY36 pKa = 9.27QHH38 pKa = 6.41EE39 pKa = 4.47VGEE42 pKa = 4.69KK43 pKa = 9.95GTDD46 pKa = 2.78HH47 pKa = 5.66YY48 pKa = 11.33QGYY51 pKa = 9.51FEE53 pKa = 5.48LLRR56 pKa = 11.84PQHH59 pKa = 6.13LSWIKK64 pKa = 10.51RR65 pKa = 11.84NISNEE70 pKa = 3.8MHH72 pKa = 7.03VEE74 pKa = 3.83MCFGDD79 pKa = 3.89RR80 pKa = 11.84KK81 pKa = 11.2DD82 pKa = 3.77NVHH85 pKa = 6.1YY86 pKa = 10.11CSKK89 pKa = 10.41PHH91 pKa = 6.96GPDD94 pKa = 3.3DD95 pKa = 3.69EE96 pKa = 6.61CYY98 pKa = 10.45DD99 pKa = 3.91EE100 pKa = 6.11CGTICDD106 pKa = 4.68CKK108 pKa = 10.38NCKK111 pKa = 9.39KK112 pKa = 10.48ARR114 pKa = 11.84KK115 pKa = 8.94APQKK119 pKa = 10.38IKK121 pKa = 10.72DD122 pKa = 3.76IIFGGDD128 pKa = 2.75WDD130 pKa = 4.41LEE132 pKa = 4.5SGKK135 pKa = 8.94RR136 pKa = 11.84TDD138 pKa = 4.76LIEE141 pKa = 4.28FKK143 pKa = 10.77KK144 pKa = 10.85DD145 pKa = 2.95IVEE148 pKa = 4.19MSLKK152 pKa = 10.15EE153 pKa = 4.03LWEE156 pKa = 3.7KK157 pKa = 10.81HH158 pKa = 4.46YY159 pKa = 11.16FLMLRR164 pKa = 11.84YY165 pKa = 9.59RR166 pKa = 11.84NMVVHH171 pKa = 6.63MKK173 pKa = 10.76NDD175 pKa = 3.1INEE178 pKa = 4.22KK179 pKa = 9.32YY180 pKa = 10.23FVGIRR185 pKa = 11.84NVILIIGRR193 pKa = 11.84SGTGKK198 pKa = 10.03SEE200 pKa = 4.2MVNKK204 pKa = 9.25EE205 pKa = 3.37HH206 pKa = 7.23SKK208 pKa = 10.43IYY210 pKa = 10.56DD211 pKa = 3.0KK212 pKa = 11.34SLNVKK217 pKa = 10.48DD218 pKa = 3.27KK219 pKa = 11.06DD220 pKa = 3.5YY221 pKa = 10.63WEE223 pKa = 4.99KK224 pKa = 11.39YY225 pKa = 10.1DD226 pKa = 3.64GEE228 pKa = 4.33EE229 pKa = 3.95IIKK232 pKa = 10.63FEE234 pKa = 4.53DD235 pKa = 3.56FTGEE239 pKa = 3.88VEE241 pKa = 4.23MNEE244 pKa = 4.41MIRR247 pKa = 11.84ILDD250 pKa = 3.67RR251 pKa = 11.84YY252 pKa = 9.82KK253 pKa = 10.59VDD255 pKa = 2.81VCKK258 pKa = 10.27RR259 pKa = 11.84YY260 pKa = 9.94VGKK263 pKa = 8.5TPFNGKK269 pKa = 9.55VIYY272 pKa = 9.19ITTNTLPEE280 pKa = 3.71EE281 pKa = 4.43WYY283 pKa = 10.63KK284 pKa = 11.44NEE286 pKa = 4.2LNVEE290 pKa = 4.38RR291 pKa = 11.84FNALYY296 pKa = 10.38RR297 pKa = 11.84RR298 pKa = 11.84FTGYY302 pKa = 10.6VVTKK306 pKa = 9.87YY307 pKa = 10.58KK308 pKa = 10.46EE309 pKa = 4.05DD310 pKa = 3.7PVVSNCVNEE319 pKa = 4.34FKK321 pKa = 11.07DD322 pKa = 4.73DD323 pKa = 3.35ILSIMGEE330 pKa = 3.92EE331 pKa = 4.55KK332 pKa = 10.73YY333 pKa = 11.28NLFKK337 pKa = 10.9VYY339 pKa = 10.3KK340 pKa = 9.15CYY342 pKa = 11.06CHH344 pKa = 6.22MCNII348 pKa = 4.42
MM1 pKa = 7.44EE2 pKa = 5.1GKK4 pKa = 9.88SIKK7 pKa = 9.6RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 10.14LFTLNKK17 pKa = 9.56PLEE20 pKa = 4.53KK21 pKa = 10.04KK22 pKa = 10.7DD23 pKa = 3.19IDD25 pKa = 3.94FNNNVVRR32 pKa = 11.84CYY34 pKa = 10.34CYY36 pKa = 9.27QHH38 pKa = 6.41EE39 pKa = 4.47VGEE42 pKa = 4.69KK43 pKa = 9.95GTDD46 pKa = 2.78HH47 pKa = 5.66YY48 pKa = 11.33QGYY51 pKa = 9.51FEE53 pKa = 5.48LLRR56 pKa = 11.84PQHH59 pKa = 6.13LSWIKK64 pKa = 10.51RR65 pKa = 11.84NISNEE70 pKa = 3.8MHH72 pKa = 7.03VEE74 pKa = 3.83MCFGDD79 pKa = 3.89RR80 pKa = 11.84KK81 pKa = 11.2DD82 pKa = 3.77NVHH85 pKa = 6.1YY86 pKa = 10.11CSKK89 pKa = 10.41PHH91 pKa = 6.96GPDD94 pKa = 3.3DD95 pKa = 3.69EE96 pKa = 6.61CYY98 pKa = 10.45DD99 pKa = 3.91EE100 pKa = 6.11CGTICDD106 pKa = 4.68CKK108 pKa = 10.38NCKK111 pKa = 9.39KK112 pKa = 10.48ARR114 pKa = 11.84KK115 pKa = 8.94APQKK119 pKa = 10.38IKK121 pKa = 10.72DD122 pKa = 3.76IIFGGDD128 pKa = 2.75WDD130 pKa = 4.41LEE132 pKa = 4.5SGKK135 pKa = 8.94RR136 pKa = 11.84TDD138 pKa = 4.76LIEE141 pKa = 4.28FKK143 pKa = 10.77KK144 pKa = 10.85DD145 pKa = 2.95IVEE148 pKa = 4.19MSLKK152 pKa = 10.15EE153 pKa = 4.03LWEE156 pKa = 3.7KK157 pKa = 10.81HH158 pKa = 4.46YY159 pKa = 11.16FLMLRR164 pKa = 11.84YY165 pKa = 9.59RR166 pKa = 11.84NMVVHH171 pKa = 6.63MKK173 pKa = 10.76NDD175 pKa = 3.1INEE178 pKa = 4.22KK179 pKa = 9.32YY180 pKa = 10.23FVGIRR185 pKa = 11.84NVILIIGRR193 pKa = 11.84SGTGKK198 pKa = 10.03SEE200 pKa = 4.2MVNKK204 pKa = 9.25EE205 pKa = 3.37HH206 pKa = 7.23SKK208 pKa = 10.43IYY210 pKa = 10.56DD211 pKa = 3.0KK212 pKa = 11.34SLNVKK217 pKa = 10.48DD218 pKa = 3.27KK219 pKa = 11.06DD220 pKa = 3.5YY221 pKa = 10.63WEE223 pKa = 4.99KK224 pKa = 11.39YY225 pKa = 10.1DD226 pKa = 3.64GEE228 pKa = 4.33EE229 pKa = 3.95IIKK232 pKa = 10.63FEE234 pKa = 4.53DD235 pKa = 3.56FTGEE239 pKa = 3.88VEE241 pKa = 4.23MNEE244 pKa = 4.41MIRR247 pKa = 11.84ILDD250 pKa = 3.67RR251 pKa = 11.84YY252 pKa = 9.82KK253 pKa = 10.59VDD255 pKa = 2.81VCKK258 pKa = 10.27RR259 pKa = 11.84YY260 pKa = 9.94VGKK263 pKa = 8.5TPFNGKK269 pKa = 9.55VIYY272 pKa = 9.19ITTNTLPEE280 pKa = 3.71EE281 pKa = 4.43WYY283 pKa = 10.63KK284 pKa = 11.44NEE286 pKa = 4.2LNVEE290 pKa = 4.38RR291 pKa = 11.84FNALYY296 pKa = 10.38RR297 pKa = 11.84RR298 pKa = 11.84FTGYY302 pKa = 10.6VVTKK306 pKa = 9.87YY307 pKa = 10.58KK308 pKa = 10.46EE309 pKa = 4.05DD310 pKa = 3.7PVVSNCVNEE319 pKa = 4.34FKK321 pKa = 11.07DD322 pKa = 4.73DD323 pKa = 3.35ILSIMGEE330 pKa = 3.92EE331 pKa = 4.55KK332 pKa = 10.73YY333 pKa = 11.28NLFKK337 pKa = 10.9VYY339 pKa = 10.3KK340 pKa = 9.15CYY342 pKa = 11.06CHH344 pKa = 6.22MCNII348 pKa = 4.42
Molecular weight: 41.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2W9F3|A0A5Q2W9F3_9CIRC ATP-dependent helicase Rep OS=Circoviridae sp. ctfvP2 OX=2656728 PE=3 SV=1
MM1 pKa = 7.36KK2 pKa = 10.34NIYY5 pKa = 9.47IYY7 pKa = 10.33KK8 pKa = 10.02RR9 pKa = 11.84KK10 pKa = 9.77NFYY13 pKa = 10.83SISFKK18 pKa = 11.18NFLLLYY24 pKa = 10.16QDD26 pKa = 3.8GKK28 pKa = 10.1MVNPTCIYY36 pKa = 9.5CNKK39 pKa = 9.5VLLYY43 pKa = 9.76LHH45 pKa = 5.99IHH47 pKa = 6.06KK48 pKa = 10.37LRR50 pKa = 11.84RR51 pKa = 11.84QKK53 pKa = 10.94PNTCICGSLLFCCHH67 pKa = 7.19CDD69 pKa = 3.41CNIYY73 pKa = 10.02IYY75 pKa = 10.53CFKK78 pKa = 10.36CCNGRR83 pKa = 11.84KK84 pKa = 9.27INN86 pKa = 3.65
MM1 pKa = 7.36KK2 pKa = 10.34NIYY5 pKa = 9.47IYY7 pKa = 10.33KK8 pKa = 10.02RR9 pKa = 11.84KK10 pKa = 9.77NFYY13 pKa = 10.83SISFKK18 pKa = 11.18NFLLLYY24 pKa = 10.16QDD26 pKa = 3.8GKK28 pKa = 10.1MVNPTCIYY36 pKa = 9.5CNKK39 pKa = 9.5VLLYY43 pKa = 9.76LHH45 pKa = 5.99IHH47 pKa = 6.06KK48 pKa = 10.37LRR50 pKa = 11.84RR51 pKa = 11.84QKK53 pKa = 10.94PNTCICGSLLFCCHH67 pKa = 7.19CDD69 pKa = 3.41CNIYY73 pKa = 10.02IYY75 pKa = 10.53CFKK78 pKa = 10.36CCNGRR83 pKa = 11.84KK84 pKa = 9.27INN86 pKa = 3.65
Molecular weight: 10.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
434 |
86 |
348 |
217.0 |
26.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
0.691 ± 0.34 | 5.76 ± 3.458 |
6.221 ± 1.916 | 7.373 ± 3.627 |
4.608 ± 0.593 | 5.3 ± 0.891 |
2.995 ± 0.242 | 7.834 ± 1.294 |
11.751 ± 0.061 | 6.682 ± 1.861 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.226 ± 0.443 | 7.604 ± 1.407 |
2.304 ± 0.011 | 1.382 ± 0.464 |
5.3 ± 0.319 | 3.456 ± 0.016 |
3.226 ± 0.443 | 5.991 ± 1.803 |
1.152 ± 0.567 | 7.143 ± 1.062 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
