
Cyphellophora europaea CBS 101466
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Chaetothyriomycetidae; Chaetothyriales; Cyphellophoraceae; Cyphellophora; Cyphellophora europaea
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
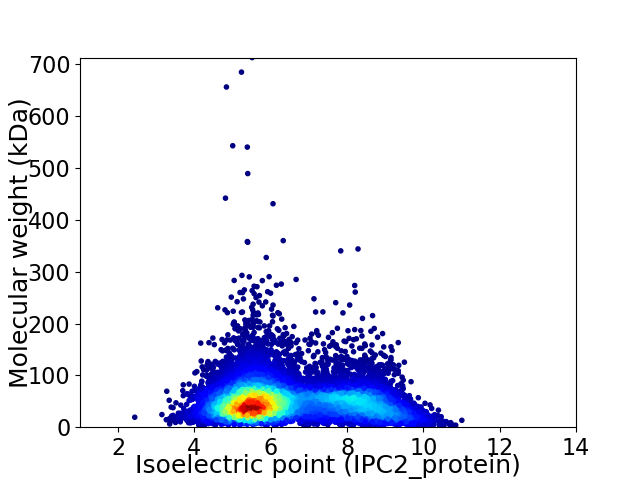
Virtual 2D-PAGE plot for 11090 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W2RLC9|W2RLC9_9EURO Uncharacterized protein OS=Cyphellophora europaea CBS 101466 OX=1220924 GN=HMPREF1541_08812 PE=4 SV=1
MM1 pKa = 7.62APGIPTAVSADD12 pKa = 3.6HH13 pKa = 6.58QPTKK17 pKa = 8.74TVRR20 pKa = 11.84IADD23 pKa = 4.15DD24 pKa = 5.01GEE26 pKa = 4.39DD27 pKa = 3.59TMQDD31 pKa = 3.2AQVSPLQSAYY41 pKa = 8.79EE42 pKa = 4.08RR43 pKa = 11.84KK44 pKa = 9.9ADD46 pKa = 3.63AVKK49 pKa = 9.26EE50 pKa = 4.11TVIRR54 pKa = 11.84GGRR57 pKa = 11.84PGVVDD62 pKa = 4.25NGSSDD67 pKa = 3.14IPSTYY72 pKa = 10.55KK73 pKa = 10.62PVDD76 pKa = 3.5EE77 pKa = 6.23DD78 pKa = 4.51DD79 pKa = 5.79DD80 pKa = 4.76SEE82 pKa = 4.32QVAPYY87 pKa = 10.38FPDD90 pKa = 3.07QDD92 pKa = 3.9ANKK95 pKa = 7.34PTTWGFHH102 pKa = 6.24ILDD105 pKa = 3.8NNQPGFAVDD114 pKa = 3.55NAVTEE119 pKa = 4.09QAEE122 pKa = 4.44DD123 pKa = 4.17GDD125 pKa = 5.17DD126 pKa = 4.68DD127 pKa = 5.13SQATPNNSLGDD138 pKa = 3.86DD139 pKa = 4.01TDD141 pKa = 3.65THH143 pKa = 6.06HH144 pKa = 7.59TDD146 pKa = 3.03AQVEE150 pKa = 4.33GGVDD154 pKa = 3.54GGDD157 pKa = 3.67GQNGDD162 pKa = 3.99QEE164 pKa = 5.52AGDD167 pKa = 4.69DD168 pKa = 4.14GQAVTDD174 pKa = 4.1SVAADD179 pKa = 3.55VAAQAAAADD188 pKa = 4.14ADD190 pKa = 4.26STQNWLVAQDD200 pKa = 3.0SWLNQLAAEE209 pKa = 4.28NPEE212 pKa = 3.78EE213 pKa = 4.07FARR216 pKa = 11.84LVEE219 pKa = 4.41SGVVDD224 pKa = 3.73PAFIPHH230 pKa = 7.15AAAQEE235 pKa = 4.37NIDD238 pKa = 3.97GDD240 pKa = 4.01GGADD244 pKa = 3.51GNTTATGAEE253 pKa = 4.16TEE255 pKa = 4.31INSDD259 pKa = 3.51EE260 pKa = 4.17AVAAGPDD267 pKa = 3.82GANDD271 pKa = 3.51SADD274 pKa = 4.18AEE276 pKa = 4.68DD277 pKa = 5.99EE278 pKa = 4.56PDD280 pKa = 3.8TDD282 pKa = 5.94DD283 pKa = 6.05NDD285 pKa = 4.17PADD288 pKa = 4.4NGNATATAPNTDD300 pKa = 3.03HH301 pKa = 7.06FVWLANRR308 pKa = 11.84NDD310 pKa = 3.59AQEE313 pKa = 4.52EE314 pKa = 4.37NTSCPACDD322 pKa = 3.34TAFSASDD329 pKa = 3.27VVIYY333 pKa = 8.65LNCGHH338 pKa = 6.31SWCSEE343 pKa = 3.94CVNLNYY349 pKa = 10.33RR350 pKa = 11.84SALRR354 pKa = 11.84SRR356 pKa = 11.84SSWPPQCCRR365 pKa = 11.84IDD367 pKa = 3.53IDD369 pKa = 3.92HH370 pKa = 7.01GSIAHH375 pKa = 6.73LLEE378 pKa = 5.17EE379 pKa = 4.91DD380 pKa = 3.63LQMDD384 pKa = 4.41LVVKK388 pKa = 10.58IEE390 pKa = 4.05EE391 pKa = 4.24FADD394 pKa = 3.63NNPVYY399 pKa = 10.61CQTPKK404 pKa = 10.63CAGGYY409 pKa = 9.35IPVEE413 pKa = 3.82RR414 pKa = 11.84RR415 pKa = 11.84EE416 pKa = 4.27GQWAGCQTCRR426 pKa = 11.84LTTCVEE432 pKa = 4.44CKK434 pKa = 10.25APAMAHH440 pKa = 6.3PMPALHH446 pKa = 7.08PNMIEE451 pKa = 4.14QVDD454 pKa = 3.77KK455 pKa = 11.17EE456 pKa = 4.13LAEE459 pKa = 4.3GQGWKK464 pKa = 9.37QCPGCRR470 pKa = 11.84NLVEE474 pKa = 4.54RR475 pKa = 11.84SGGCNYY481 pKa = 9.16MSCEE485 pKa = 4.37CGHH488 pKa = 6.45HH489 pKa = 6.52FCYY492 pKa = 10.28QCGGTLEE499 pKa = 4.96NNMPCDD505 pKa = 4.93CEE507 pKa = 4.91GQQPWVAQMNQEE519 pKa = 4.08AAGGADD525 pKa = 3.13QDD527 pKa = 4.05GDD529 pKa = 4.09SEE531 pKa = 4.78DD532 pKa = 4.13EE533 pKa = 4.09EE534 pKa = 6.58GEE536 pKa = 4.21DD537 pKa = 4.65GEE539 pKa = 5.08RR540 pKa = 11.84DD541 pKa = 3.56EE542 pKa = 6.14NEE544 pKa = 4.92DD545 pKa = 3.43GTEE548 pKa = 4.71DD549 pKa = 3.98EE550 pKa = 6.16AGDD553 pKa = 3.66QDD555 pKa = 4.88GDD557 pKa = 3.94EE558 pKa = 4.26QVQPAVQDD566 pKa = 3.62GGATGGWW573 pKa = 3.38
MM1 pKa = 7.62APGIPTAVSADD12 pKa = 3.6HH13 pKa = 6.58QPTKK17 pKa = 8.74TVRR20 pKa = 11.84IADD23 pKa = 4.15DD24 pKa = 5.01GEE26 pKa = 4.39DD27 pKa = 3.59TMQDD31 pKa = 3.2AQVSPLQSAYY41 pKa = 8.79EE42 pKa = 4.08RR43 pKa = 11.84KK44 pKa = 9.9ADD46 pKa = 3.63AVKK49 pKa = 9.26EE50 pKa = 4.11TVIRR54 pKa = 11.84GGRR57 pKa = 11.84PGVVDD62 pKa = 4.25NGSSDD67 pKa = 3.14IPSTYY72 pKa = 10.55KK73 pKa = 10.62PVDD76 pKa = 3.5EE77 pKa = 6.23DD78 pKa = 4.51DD79 pKa = 5.79DD80 pKa = 4.76SEE82 pKa = 4.32QVAPYY87 pKa = 10.38FPDD90 pKa = 3.07QDD92 pKa = 3.9ANKK95 pKa = 7.34PTTWGFHH102 pKa = 6.24ILDD105 pKa = 3.8NNQPGFAVDD114 pKa = 3.55NAVTEE119 pKa = 4.09QAEE122 pKa = 4.44DD123 pKa = 4.17GDD125 pKa = 5.17DD126 pKa = 4.68DD127 pKa = 5.13SQATPNNSLGDD138 pKa = 3.86DD139 pKa = 4.01TDD141 pKa = 3.65THH143 pKa = 6.06HH144 pKa = 7.59TDD146 pKa = 3.03AQVEE150 pKa = 4.33GGVDD154 pKa = 3.54GGDD157 pKa = 3.67GQNGDD162 pKa = 3.99QEE164 pKa = 5.52AGDD167 pKa = 4.69DD168 pKa = 4.14GQAVTDD174 pKa = 4.1SVAADD179 pKa = 3.55VAAQAAAADD188 pKa = 4.14ADD190 pKa = 4.26STQNWLVAQDD200 pKa = 3.0SWLNQLAAEE209 pKa = 4.28NPEE212 pKa = 3.78EE213 pKa = 4.07FARR216 pKa = 11.84LVEE219 pKa = 4.41SGVVDD224 pKa = 3.73PAFIPHH230 pKa = 7.15AAAQEE235 pKa = 4.37NIDD238 pKa = 3.97GDD240 pKa = 4.01GGADD244 pKa = 3.51GNTTATGAEE253 pKa = 4.16TEE255 pKa = 4.31INSDD259 pKa = 3.51EE260 pKa = 4.17AVAAGPDD267 pKa = 3.82GANDD271 pKa = 3.51SADD274 pKa = 4.18AEE276 pKa = 4.68DD277 pKa = 5.99EE278 pKa = 4.56PDD280 pKa = 3.8TDD282 pKa = 5.94DD283 pKa = 6.05NDD285 pKa = 4.17PADD288 pKa = 4.4NGNATATAPNTDD300 pKa = 3.03HH301 pKa = 7.06FVWLANRR308 pKa = 11.84NDD310 pKa = 3.59AQEE313 pKa = 4.52EE314 pKa = 4.37NTSCPACDD322 pKa = 3.34TAFSASDD329 pKa = 3.27VVIYY333 pKa = 8.65LNCGHH338 pKa = 6.31SWCSEE343 pKa = 3.94CVNLNYY349 pKa = 10.33RR350 pKa = 11.84SALRR354 pKa = 11.84SRR356 pKa = 11.84SSWPPQCCRR365 pKa = 11.84IDD367 pKa = 3.53IDD369 pKa = 3.92HH370 pKa = 7.01GSIAHH375 pKa = 6.73LLEE378 pKa = 5.17EE379 pKa = 4.91DD380 pKa = 3.63LQMDD384 pKa = 4.41LVVKK388 pKa = 10.58IEE390 pKa = 4.05EE391 pKa = 4.24FADD394 pKa = 3.63NNPVYY399 pKa = 10.61CQTPKK404 pKa = 10.63CAGGYY409 pKa = 9.35IPVEE413 pKa = 3.82RR414 pKa = 11.84RR415 pKa = 11.84EE416 pKa = 4.27GQWAGCQTCRR426 pKa = 11.84LTTCVEE432 pKa = 4.44CKK434 pKa = 10.25APAMAHH440 pKa = 6.3PMPALHH446 pKa = 7.08PNMIEE451 pKa = 4.14QVDD454 pKa = 3.77KK455 pKa = 11.17EE456 pKa = 4.13LAEE459 pKa = 4.3GQGWKK464 pKa = 9.37QCPGCRR470 pKa = 11.84NLVEE474 pKa = 4.54RR475 pKa = 11.84SGGCNYY481 pKa = 9.16MSCEE485 pKa = 4.37CGHH488 pKa = 6.45HH489 pKa = 6.52FCYY492 pKa = 10.28QCGGTLEE499 pKa = 4.96NNMPCDD505 pKa = 4.93CEE507 pKa = 4.91GQQPWVAQMNQEE519 pKa = 4.08AAGGADD525 pKa = 3.13QDD527 pKa = 4.05GDD529 pKa = 4.09SEE531 pKa = 4.78DD532 pKa = 4.13EE533 pKa = 4.09EE534 pKa = 6.58GEE536 pKa = 4.21DD537 pKa = 4.65GEE539 pKa = 5.08RR540 pKa = 11.84DD541 pKa = 3.56EE542 pKa = 6.14NEE544 pKa = 4.92DD545 pKa = 3.43GTEE548 pKa = 4.71DD549 pKa = 3.98EE550 pKa = 6.16AGDD553 pKa = 3.66QDD555 pKa = 4.88GDD557 pKa = 3.94EE558 pKa = 4.26QVQPAVQDD566 pKa = 3.62GGATGGWW573 pKa = 3.38
Molecular weight: 60.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W2RJT9|W2RJT9_9EURO Uncharacterized protein OS=Cyphellophora europaea CBS 101466 OX=1220924 GN=HMPREF1541_09030 PE=4 SV=1
MM1 pKa = 7.41FGTRR5 pKa = 11.84RR6 pKa = 11.84SPRR9 pKa = 11.84THH11 pKa = 6.84AAPRR15 pKa = 11.84STRR18 pKa = 11.84HH19 pKa = 4.5TTTTTKK25 pKa = 10.14PGIMSRR31 pKa = 11.84LTGGGRR37 pKa = 11.84TRR39 pKa = 11.84KK40 pKa = 7.3THH42 pKa = 4.68TTTTTTAPRR51 pKa = 11.84TQRR54 pKa = 11.84KK55 pKa = 9.24SGWGTRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84APRR66 pKa = 11.84AAPVATHH73 pKa = 6.27HH74 pKa = 6.29RR75 pKa = 11.84RR76 pKa = 11.84KK77 pKa = 9.93PSMGDD82 pKa = 3.11KK83 pKa = 10.62VAGALKK89 pKa = 10.14KK90 pKa = 10.61VQGSLTRR97 pKa = 11.84NPAKK101 pKa = 10.27KK102 pKa = 10.0AAGTRR107 pKa = 11.84RR108 pKa = 11.84MHH110 pKa = 5.68GTDD113 pKa = 2.87GRR115 pKa = 11.84GSRR118 pKa = 11.84RR119 pKa = 11.84VYY121 pKa = 10.87
MM1 pKa = 7.41FGTRR5 pKa = 11.84RR6 pKa = 11.84SPRR9 pKa = 11.84THH11 pKa = 6.84AAPRR15 pKa = 11.84STRR18 pKa = 11.84HH19 pKa = 4.5TTTTTKK25 pKa = 10.14PGIMSRR31 pKa = 11.84LTGGGRR37 pKa = 11.84TRR39 pKa = 11.84KK40 pKa = 7.3THH42 pKa = 4.68TTTTTTAPRR51 pKa = 11.84TQRR54 pKa = 11.84KK55 pKa = 9.24SGWGTRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84APRR66 pKa = 11.84AAPVATHH73 pKa = 6.27HH74 pKa = 6.29RR75 pKa = 11.84RR76 pKa = 11.84KK77 pKa = 9.93PSMGDD82 pKa = 3.11KK83 pKa = 10.62VAGALKK89 pKa = 10.14KK90 pKa = 10.61VQGSLTRR97 pKa = 11.84NPAKK101 pKa = 10.27KK102 pKa = 10.0AAGTRR107 pKa = 11.84RR108 pKa = 11.84MHH110 pKa = 5.68GTDD113 pKa = 2.87GRR115 pKa = 11.84GSRR118 pKa = 11.84RR119 pKa = 11.84VYY121 pKa = 10.87
Molecular weight: 13.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5402066 |
30 |
6474 |
487.1 |
53.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.093 ± 0.02 | 1.153 ± 0.008 |
5.775 ± 0.017 | 6.064 ± 0.018 |
3.616 ± 0.012 | 7.07 ± 0.019 |
2.402 ± 0.01 | 4.641 ± 0.014 |
4.677 ± 0.02 | 8.863 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.209 ± 0.007 | 3.544 ± 0.012 |
6.041 ± 0.02 | 4.164 ± 0.017 |
6.18 ± 0.018 | 8.073 ± 0.023 |
6.002 ± 0.016 | 6.205 ± 0.016 |
1.497 ± 0.009 | 2.73 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
