
Antricoccus suffuscus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Antricoccaceae; Antricoccus
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
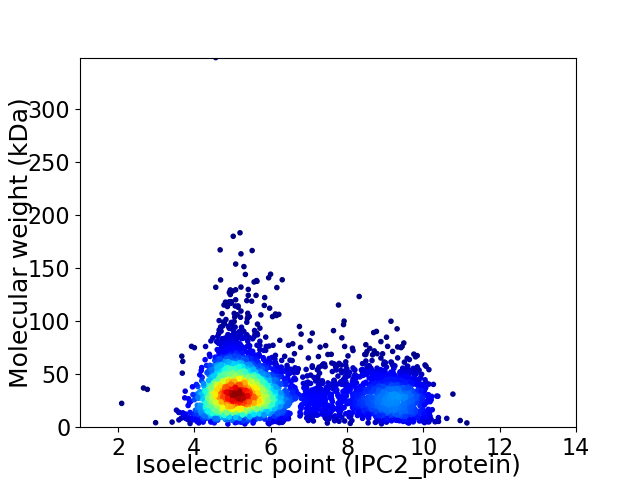
Virtual 2D-PAGE plot for 4340 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T1A0B2|A0A2T1A0B2_9ACTN Stearoyl-CoA desaturase (Delta-9 desaturase) OS=Antricoccus suffuscus OX=1629062 GN=CLV47_10753 PE=4 SV=1
MM1 pKa = 7.67AAPITFDD8 pKa = 3.54SLVGYY13 pKa = 10.49AIANPLAICEE23 pKa = 4.45SVLVANPVEE32 pKa = 4.51VEE34 pKa = 3.65ALGAVFRR41 pKa = 11.84AAGASTAEE49 pKa = 4.2SAMCALEE56 pKa = 4.03AAQLADD62 pKa = 3.8AAGMIDD68 pKa = 5.29DD69 pKa = 4.32KK70 pKa = 11.66TPVQLMAEE78 pKa = 4.29VTATKK83 pKa = 9.29DD84 pKa = 3.36TLDD87 pKa = 3.66VNSAQLDD94 pKa = 4.4LIGGLLSEE102 pKa = 4.88VGSQIHH108 pKa = 6.7LVQAKK113 pKa = 10.09LNSLLDD119 pKa = 3.38MMYY122 pKa = 11.19AEE124 pKa = 4.96IATAASMYY132 pKa = 9.64EE133 pKa = 4.12CAVPYY138 pKa = 10.72DD139 pKa = 4.02PANDD143 pKa = 3.8AALHH147 pKa = 5.22EE148 pKa = 4.49QCAQAALASINTTATSVNAVVDD170 pKa = 4.62GYY172 pKa = 11.42DD173 pKa = 4.06AFLTDD178 pKa = 5.21HH179 pKa = 7.24LATLQEE185 pKa = 4.65DD186 pKa = 4.2YY187 pKa = 11.31GYY189 pKa = 11.14VPPAALNDD197 pKa = 3.94GAALGNDD204 pKa = 3.72PAYY207 pKa = 11.1VSLTTGMPGASAPPGQVQQWWNEE230 pKa = 3.15RR231 pKa = 11.84SAAEE235 pKa = 4.23KK236 pKa = 10.51VWLIDD241 pKa = 3.5NEE243 pKa = 4.38GAALSMMHH251 pKa = 7.02GLPALVLNLVNRR263 pKa = 11.84RR264 pKa = 11.84EE265 pKa = 4.23LTDD268 pKa = 3.96DD269 pKa = 3.88ASSIDD274 pKa = 3.52AQVAMVEE281 pKa = 4.31TQKK284 pKa = 11.42ASLMQILGVSVASDD298 pKa = 3.62VFDD301 pKa = 4.51HH302 pKa = 7.09PDD304 pKa = 2.87IRR306 pKa = 11.84ASNPGAADD314 pKa = 3.43EE315 pKa = 4.43LAGVLPVLAALKK327 pKa = 10.04VKK329 pKa = 9.75QANIHH334 pKa = 4.96GVEE337 pKa = 4.53DD338 pKa = 3.87GAKK341 pKa = 9.31AVAMADD347 pKa = 3.16GTAIKK352 pKa = 9.52TYY354 pKa = 10.84LLDD357 pKa = 4.1YY358 pKa = 10.85DD359 pKa = 4.89DD360 pKa = 6.02LGSADD365 pKa = 4.44DD366 pKa = 4.45GEE368 pKa = 4.37AVIAIGNPDD377 pKa = 3.73EE378 pKa = 4.58AADD381 pKa = 3.51IAVIVPGTTSSARR394 pKa = 11.84TMSDD398 pKa = 3.42YY399 pKa = 11.3VGSGGNLYY407 pKa = 11.06AEE409 pKa = 4.76MNDD412 pKa = 3.33VDD414 pKa = 4.51PDD416 pKa = 3.92SKK418 pKa = 11.2KK419 pKa = 11.11SVIAYY424 pKa = 10.26LGMDD428 pKa = 4.48APDD431 pKa = 4.07TLPDD435 pKa = 3.6ASLPIYY441 pKa = 10.63ANEE444 pKa = 4.22AAPEE448 pKa = 4.01LAADD452 pKa = 3.28IAGYY456 pKa = 8.28TEE458 pKa = 4.61SNLVATGADD467 pKa = 3.25PHH469 pKa = 5.75VTVIGHH475 pKa = 6.06SYY477 pKa = 10.38GSLVVGAALEE487 pKa = 4.43SGDD490 pKa = 5.48LIADD494 pKa = 3.55DD495 pKa = 4.98VIFVGSPGVGVKK507 pKa = 10.36NVDD510 pKa = 3.96EE511 pKa = 4.91LNMDD515 pKa = 3.97GSHH518 pKa = 6.33VYY520 pKa = 10.54VGLLPDD526 pKa = 4.72DD527 pKa = 5.54AISNANDD534 pKa = 3.4FFDD537 pKa = 3.87MKK539 pKa = 10.71AGSTLGVPDD548 pKa = 4.4GAWFGHH554 pKa = 5.37NPARR558 pKa = 11.84EE559 pKa = 4.33SFGGTVFDD567 pKa = 4.3TDD569 pKa = 3.09TDD571 pKa = 4.17GDD573 pKa = 3.74HH574 pKa = 7.3GDD576 pKa = 3.64YY577 pKa = 10.62FDD579 pKa = 5.36YY580 pKa = 9.1GTTQLDD586 pKa = 3.24NMAAVATGQYY596 pKa = 11.15DD597 pKa = 4.32EE598 pKa = 4.66VTLDD602 pKa = 3.21
MM1 pKa = 7.67AAPITFDD8 pKa = 3.54SLVGYY13 pKa = 10.49AIANPLAICEE23 pKa = 4.45SVLVANPVEE32 pKa = 4.51VEE34 pKa = 3.65ALGAVFRR41 pKa = 11.84AAGASTAEE49 pKa = 4.2SAMCALEE56 pKa = 4.03AAQLADD62 pKa = 3.8AAGMIDD68 pKa = 5.29DD69 pKa = 4.32KK70 pKa = 11.66TPVQLMAEE78 pKa = 4.29VTATKK83 pKa = 9.29DD84 pKa = 3.36TLDD87 pKa = 3.66VNSAQLDD94 pKa = 4.4LIGGLLSEE102 pKa = 4.88VGSQIHH108 pKa = 6.7LVQAKK113 pKa = 10.09LNSLLDD119 pKa = 3.38MMYY122 pKa = 11.19AEE124 pKa = 4.96IATAASMYY132 pKa = 9.64EE133 pKa = 4.12CAVPYY138 pKa = 10.72DD139 pKa = 4.02PANDD143 pKa = 3.8AALHH147 pKa = 5.22EE148 pKa = 4.49QCAQAALASINTTATSVNAVVDD170 pKa = 4.62GYY172 pKa = 11.42DD173 pKa = 4.06AFLTDD178 pKa = 5.21HH179 pKa = 7.24LATLQEE185 pKa = 4.65DD186 pKa = 4.2YY187 pKa = 11.31GYY189 pKa = 11.14VPPAALNDD197 pKa = 3.94GAALGNDD204 pKa = 3.72PAYY207 pKa = 11.1VSLTTGMPGASAPPGQVQQWWNEE230 pKa = 3.15RR231 pKa = 11.84SAAEE235 pKa = 4.23KK236 pKa = 10.51VWLIDD241 pKa = 3.5NEE243 pKa = 4.38GAALSMMHH251 pKa = 7.02GLPALVLNLVNRR263 pKa = 11.84RR264 pKa = 11.84EE265 pKa = 4.23LTDD268 pKa = 3.96DD269 pKa = 3.88ASSIDD274 pKa = 3.52AQVAMVEE281 pKa = 4.31TQKK284 pKa = 11.42ASLMQILGVSVASDD298 pKa = 3.62VFDD301 pKa = 4.51HH302 pKa = 7.09PDD304 pKa = 2.87IRR306 pKa = 11.84ASNPGAADD314 pKa = 3.43EE315 pKa = 4.43LAGVLPVLAALKK327 pKa = 10.04VKK329 pKa = 9.75QANIHH334 pKa = 4.96GVEE337 pKa = 4.53DD338 pKa = 3.87GAKK341 pKa = 9.31AVAMADD347 pKa = 3.16GTAIKK352 pKa = 9.52TYY354 pKa = 10.84LLDD357 pKa = 4.1YY358 pKa = 10.85DD359 pKa = 4.89DD360 pKa = 6.02LGSADD365 pKa = 4.44DD366 pKa = 4.45GEE368 pKa = 4.37AVIAIGNPDD377 pKa = 3.73EE378 pKa = 4.58AADD381 pKa = 3.51IAVIVPGTTSSARR394 pKa = 11.84TMSDD398 pKa = 3.42YY399 pKa = 11.3VGSGGNLYY407 pKa = 11.06AEE409 pKa = 4.76MNDD412 pKa = 3.33VDD414 pKa = 4.51PDD416 pKa = 3.92SKK418 pKa = 11.2KK419 pKa = 11.11SVIAYY424 pKa = 10.26LGMDD428 pKa = 4.48APDD431 pKa = 4.07TLPDD435 pKa = 3.6ASLPIYY441 pKa = 10.63ANEE444 pKa = 4.22AAPEE448 pKa = 4.01LAADD452 pKa = 3.28IAGYY456 pKa = 8.28TEE458 pKa = 4.61SNLVATGADD467 pKa = 3.25PHH469 pKa = 5.75VTVIGHH475 pKa = 6.06SYY477 pKa = 10.38GSLVVGAALEE487 pKa = 4.43SGDD490 pKa = 5.48LIADD494 pKa = 3.55DD495 pKa = 4.98VIFVGSPGVGVKK507 pKa = 10.36NVDD510 pKa = 3.96EE511 pKa = 4.91LNMDD515 pKa = 3.97GSHH518 pKa = 6.33VYY520 pKa = 10.54VGLLPDD526 pKa = 4.72DD527 pKa = 5.54AISNANDD534 pKa = 3.4FFDD537 pKa = 3.87MKK539 pKa = 10.71AGSTLGVPDD548 pKa = 4.4GAWFGHH554 pKa = 5.37NPARR558 pKa = 11.84EE559 pKa = 4.33SFGGTVFDD567 pKa = 4.3TDD569 pKa = 3.09TDD571 pKa = 4.17GDD573 pKa = 3.74HH574 pKa = 7.3GDD576 pKa = 3.64YY577 pKa = 10.62FDD579 pKa = 5.36YY580 pKa = 9.1GTTQLDD586 pKa = 3.24NMAAVATGQYY596 pKa = 11.15DD597 pKa = 4.32EE598 pKa = 4.66VTLDD602 pKa = 3.21
Molecular weight: 62.15 kDa
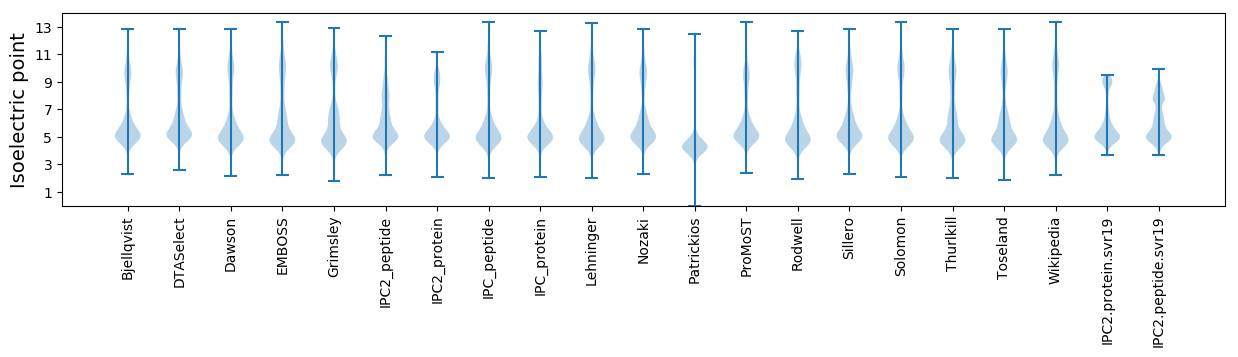
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T0ZRV4|A0A2T0ZRV4_9ACTN o-succinylbenzoate synthase OS=Antricoccus suffuscus OX=1629062 GN=menC PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.72HH17 pKa = 5.74RR18 pKa = 11.84KK19 pKa = 7.77LLKK22 pKa = 8.07RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.72HH17 pKa = 5.74RR18 pKa = 11.84KK19 pKa = 7.77LLKK22 pKa = 8.07RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
Molecular weight: 4.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1423240 |
26 |
3336 |
327.9 |
35.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.431 ± 0.039 | 0.768 ± 0.01 |
6.374 ± 0.031 | 5.253 ± 0.036 |
2.955 ± 0.022 | 8.846 ± 0.035 |
2.103 ± 0.017 | 4.906 ± 0.026 |
2.802 ± 0.028 | 9.636 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.146 ± 0.014 | 2.294 ± 0.018 |
5.374 ± 0.026 | 3.065 ± 0.02 |
6.822 ± 0.041 | 5.995 ± 0.029 |
6.152 ± 0.028 | 8.48 ± 0.03 |
1.36 ± 0.015 | 2.239 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
