
Sphingobium japonicum (strain DSM 16413 / CCM 7287 / MTCC 6362 / UT26 / NBRC 101211 / UT26S)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingobium; Sphingobium indicum
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
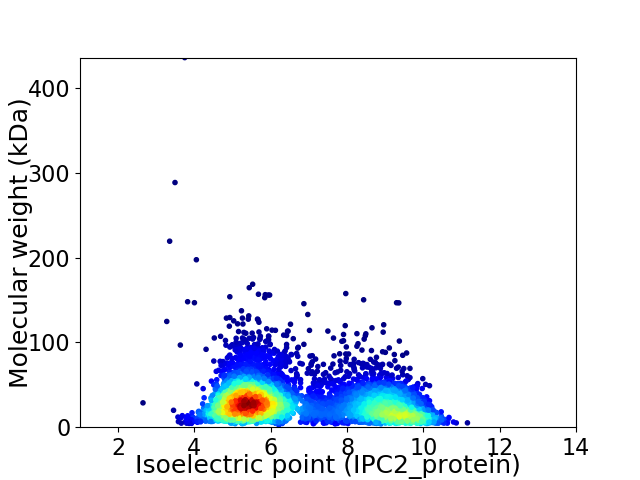
Virtual 2D-PAGE plot for 4347 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D4Z202|D4Z202_SPHJU TrbN/BfpH-like protein OS=Sphingobium japonicum (strain DSM 16413 / CCM 7287 / MTCC 6362 / UT26 / NBRC 101211 / UT26S) OX=452662 GN=SJA_C1-18000 PE=4 SV=1
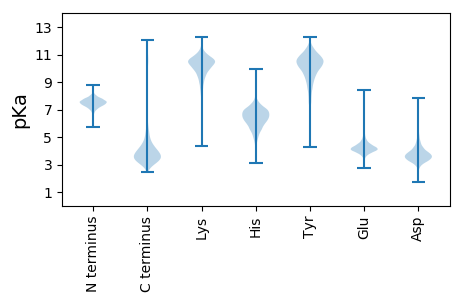
MM1 pKa = 7.7TYY3 pKa = 10.26GHH5 pKa = 6.55SVTIDD10 pKa = 3.11GSLDD14 pKa = 2.97DD15 pKa = 4.41WFATDD20 pKa = 5.81RR21 pKa = 11.84IDD23 pKa = 5.21DD24 pKa = 3.89NSAPGYY30 pKa = 9.28QVYY33 pKa = 7.91ATLDD37 pKa = 3.03NDD39 pKa = 3.86AFVFALTAPVAIGADD54 pKa = 3.38TTVWLNTDD62 pKa = 3.62GKK64 pKa = 9.32TSTGYY69 pKa = 10.64QIFGFAGGAEE79 pKa = 4.26YY80 pKa = 10.84NVNFAADD87 pKa = 3.84GTVNLYY93 pKa = 10.61SGAAGEE99 pKa = 4.19TLVLGGLQAAWSADD113 pKa = 3.2RR114 pKa = 11.84TTVEE118 pKa = 3.96FRR120 pKa = 11.84LPTSAVGNPAAMYY133 pKa = 10.12SYY135 pKa = 11.65YY136 pKa = 10.69DD137 pKa = 3.56INNLSNSFMPSNYY150 pKa = 8.91QSAPFTVFNQTTATQAPDD168 pKa = 3.24MRR170 pKa = 11.84IGIVFSQTTADD181 pKa = 3.82AYY183 pKa = 10.59FSPTAYY189 pKa = 9.99SQLFMSVQAQAIQAGIPFDD208 pKa = 3.74ILTEE212 pKa = 4.37ADD214 pKa = 3.6LTDD217 pKa = 4.26LNKK220 pKa = 10.43LSHH223 pKa = 6.51YY224 pKa = 9.61DD225 pKa = 3.36ALVFPSFRR233 pKa = 11.84NVPVDD238 pKa = 3.22KK239 pKa = 11.29VEE241 pKa = 5.14AISHH245 pKa = 4.93TLEE248 pKa = 4.06MASKK252 pKa = 10.13QLGIGLITAGEE263 pKa = 4.14FMTNQPHH270 pKa = 5.64VPGGAIGGDD279 pKa = 4.09PLPGDD284 pKa = 4.91PYY286 pKa = 11.72AQMKK290 pKa = 10.45LLFDD294 pKa = 3.87ATRR297 pKa = 11.84VTGGTGDD304 pKa = 3.84VVVTATDD311 pKa = 3.56PTGLVLDD318 pKa = 5.23HH319 pKa = 6.54YY320 pKa = 11.91ANGQQVNSYY329 pKa = 11.03SGVGWNAFASVSGTGHH345 pKa = 6.44QIATEE350 pKa = 4.42TINGGSAYY358 pKa = 10.08AAALATQTGAARR370 pKa = 11.84NVLFSSEE377 pKa = 3.75GVMADD382 pKa = 4.33ANMLQQAISYY392 pKa = 8.11AANGTGLSVGLHH404 pKa = 6.58LSRR407 pKa = 11.84EE408 pKa = 4.38SGVVAARR415 pKa = 11.84VDD417 pKa = 3.45MDD419 pKa = 3.35QSQEE423 pKa = 4.17LEE425 pKa = 4.4DD426 pKa = 3.84VDD428 pKa = 5.0PPGSTPGIYY437 pKa = 10.21DD438 pKa = 3.48KK439 pKa = 11.03LLPILTQWKK448 pKa = 9.71ADD450 pKa = 3.66YY451 pKa = 10.88NFVGSFYY458 pKa = 11.03LNVGNGTNGTGTDD471 pKa = 3.38WAVSLPYY478 pKa = 10.58YY479 pKa = 10.32KK480 pKa = 10.85ALLDD484 pKa = 4.35LGNEE488 pKa = 4.38LGTHH492 pKa = 6.09SLTHH496 pKa = 7.0PDD498 pKa = 3.09NTNLLNAQQIQTEE511 pKa = 4.41FQDD514 pKa = 4.37SIAILNQQISAYY526 pKa = 10.08LNQPFQITGAAVPGAPEE543 pKa = 4.11SLPVSQEE550 pKa = 3.05IMKK553 pKa = 10.38YY554 pKa = 10.05VSDD557 pKa = 4.24YY558 pKa = 10.76LTGGYY563 pKa = 9.32AAQGAGYY570 pKa = 8.77PNAFGYY576 pKa = 7.41LTPADD581 pKa = 3.35QSKK584 pKa = 10.57IYY586 pKa = 9.4FAPNTSFDD594 pKa = 3.39YY595 pKa = 11.03TFIEE599 pKa = 4.92FQQKK603 pKa = 7.0TVAEE607 pKa = 4.48ATALWIGEE615 pKa = 3.92FDD617 pKa = 4.14KK618 pKa = 11.48LVANAGMPVIVWPWHH633 pKa = 6.94DD634 pKa = 3.57YY635 pKa = 10.36GAADD639 pKa = 4.58FLSGSPTVDD648 pKa = 2.81SPYY651 pKa = 10.57DD652 pKa = 3.35ISLFTSWIQHH662 pKa = 4.4AVAANMEE669 pKa = 4.58FVTLDD674 pKa = 3.39DD675 pKa = 3.79LAARR679 pKa = 11.84MTAVKK684 pKa = 10.21NATITTTVAGNSITASVGGANVGNLSLDD712 pKa = 3.64VEE714 pKa = 4.49RR715 pKa = 11.84QGALVIQNVTGWYY728 pKa = 9.97AYY730 pKa = 10.57DD731 pKa = 3.64SDD733 pKa = 5.55SVFLPQSGGTFKK745 pKa = 9.84ITLGAAADD753 pKa = 4.2DD754 pKa = 4.07VTHH757 pKa = 6.31ITDD760 pKa = 3.66LPMRR764 pKa = 11.84AVLLSVAGDD773 pKa = 3.42GHH775 pKa = 6.51NLAFSIQGEE784 pKa = 4.55GKK786 pKa = 10.33VVIDD790 pKa = 3.9IANPGTDD797 pKa = 3.75GIVVTGAAGYY807 pKa = 8.64SQVGEE812 pKa = 4.24IVTVDD817 pKa = 3.19LGGYY821 pKa = 8.58GLHH824 pKa = 6.52NVMLEE829 pKa = 4.09YY830 pKa = 10.67KK831 pKa = 9.21NPPVIVSNGGGPTATLSMGEE851 pKa = 4.19NGTAVTTVVATDD863 pKa = 3.59ADD865 pKa = 3.83VGTVIGYY872 pKa = 9.33SISGGADD879 pKa = 2.87AAKK882 pKa = 10.28FRR884 pKa = 11.84IDD886 pKa = 3.38GATGALTFLSAPNFEE901 pKa = 4.5AAGDD905 pKa = 3.74VGTDD909 pKa = 3.01NVYY912 pKa = 11.12DD913 pKa = 4.11VIVRR917 pKa = 11.84ASDD920 pKa = 3.12GTLYY924 pKa = 10.68DD925 pKa = 3.86DD926 pKa = 3.62QAIAVTITNVNEE938 pKa = 4.03APVITSNGGGASAAVSVAEE957 pKa = 4.16NLTGVTTVTAIDD969 pKa = 4.0PDD971 pKa = 4.25AGTMLAYY978 pKa = 10.45SIVGGPDD985 pKa = 2.74RR986 pKa = 11.84ALFSIDD992 pKa = 3.59PSTGALAFLSPPNYY1006 pKa = 10.02DD1007 pKa = 3.99LPGDD1011 pKa = 3.83ANGNNVYY1018 pKa = 10.62SLTVRR1023 pKa = 11.84VSDD1026 pKa = 3.91GALFDD1031 pKa = 4.18DD1032 pKa = 3.92QVINVTVTNVNEE1044 pKa = 3.96QAPVITSGGGGDD1056 pKa = 3.39TAAISVSSGTRR1067 pKa = 11.84TVMKK1071 pKa = 9.27VTAVDD1076 pKa = 3.51PDD1078 pKa = 4.0ANTTLAYY1085 pKa = 10.36SIAGGVDD1092 pKa = 2.76AGKK1095 pKa = 8.57FTINASTGQLNFVSTPNYY1113 pKa = 10.27LLPTDD1118 pKa = 4.13AGRR1121 pKa = 11.84DD1122 pKa = 3.39NVYY1125 pKa = 10.97DD1126 pKa = 4.04VIVRR1130 pKa = 11.84ASDD1133 pKa = 3.47GTLHH1137 pKa = 7.35DD1138 pKa = 4.0EE1139 pKa = 3.91QAIAVTVTILNAPPIIISNGGGASATIRR1167 pKa = 11.84MAEE1170 pKa = 4.19NGTAVTKK1177 pKa = 9.35VTATDD1182 pKa = 3.41VNLGQSITYY1191 pKa = 10.06SLGSAVDD1198 pKa = 3.7GALFSIDD1205 pKa = 3.73PQSGVLTFKK1214 pKa = 10.53SAPDD1218 pKa = 3.74FEE1220 pKa = 6.3APADD1224 pKa = 3.87SNLDD1228 pKa = 2.83NRR1230 pKa = 11.84YY1231 pKa = 8.85QVQVLATDD1239 pKa = 3.44SLGAQDD1245 pKa = 4.31FQNLTVVVNNVSGLSLTAASGGSTLIGQGEE1275 pKa = 4.67EE1276 pKa = 3.94DD1277 pKa = 3.82TLRR1280 pKa = 11.84GAGGNDD1286 pKa = 3.26TLIGNGGSDD1295 pKa = 3.73TLNGAGGNDD1304 pKa = 4.11NIDD1307 pKa = 3.63GGAGADD1313 pKa = 4.32RR1314 pKa = 11.84ITGGAGADD1322 pKa = 3.5ILTGGADD1329 pKa = 3.7ADD1331 pKa = 4.0LFIYY1335 pKa = 10.42NATGNSTLAVMDD1347 pKa = 5.38RR1348 pKa = 11.84IMDD1351 pKa = 4.11FQQGIDD1357 pKa = 3.95RR1358 pKa = 11.84IDD1360 pKa = 3.57LSGIDD1365 pKa = 4.14ALSALSGNQAFSFIGNAAFSGAGQLHH1391 pKa = 6.42VYY1393 pKa = 10.97SNGTDD1398 pKa = 3.18SFVEE1402 pKa = 4.04ANVDD1406 pKa = 3.7NNPATVDD1413 pKa = 3.7FAIRR1417 pKa = 11.84LIGVHH1422 pKa = 6.35NLTASDD1428 pKa = 3.96FLLL1431 pKa = 4.74
MM1 pKa = 7.7TYY3 pKa = 10.26GHH5 pKa = 6.55SVTIDD10 pKa = 3.11GSLDD14 pKa = 2.97DD15 pKa = 4.41WFATDD20 pKa = 5.81RR21 pKa = 11.84IDD23 pKa = 5.21DD24 pKa = 3.89NSAPGYY30 pKa = 9.28QVYY33 pKa = 7.91ATLDD37 pKa = 3.03NDD39 pKa = 3.86AFVFALTAPVAIGADD54 pKa = 3.38TTVWLNTDD62 pKa = 3.62GKK64 pKa = 9.32TSTGYY69 pKa = 10.64QIFGFAGGAEE79 pKa = 4.26YY80 pKa = 10.84NVNFAADD87 pKa = 3.84GTVNLYY93 pKa = 10.61SGAAGEE99 pKa = 4.19TLVLGGLQAAWSADD113 pKa = 3.2RR114 pKa = 11.84TTVEE118 pKa = 3.96FRR120 pKa = 11.84LPTSAVGNPAAMYY133 pKa = 10.12SYY135 pKa = 11.65YY136 pKa = 10.69DD137 pKa = 3.56INNLSNSFMPSNYY150 pKa = 8.91QSAPFTVFNQTTATQAPDD168 pKa = 3.24MRR170 pKa = 11.84IGIVFSQTTADD181 pKa = 3.82AYY183 pKa = 10.59FSPTAYY189 pKa = 9.99SQLFMSVQAQAIQAGIPFDD208 pKa = 3.74ILTEE212 pKa = 4.37ADD214 pKa = 3.6LTDD217 pKa = 4.26LNKK220 pKa = 10.43LSHH223 pKa = 6.51YY224 pKa = 9.61DD225 pKa = 3.36ALVFPSFRR233 pKa = 11.84NVPVDD238 pKa = 3.22KK239 pKa = 11.29VEE241 pKa = 5.14AISHH245 pKa = 4.93TLEE248 pKa = 4.06MASKK252 pKa = 10.13QLGIGLITAGEE263 pKa = 4.14FMTNQPHH270 pKa = 5.64VPGGAIGGDD279 pKa = 4.09PLPGDD284 pKa = 4.91PYY286 pKa = 11.72AQMKK290 pKa = 10.45LLFDD294 pKa = 3.87ATRR297 pKa = 11.84VTGGTGDD304 pKa = 3.84VVVTATDD311 pKa = 3.56PTGLVLDD318 pKa = 5.23HH319 pKa = 6.54YY320 pKa = 11.91ANGQQVNSYY329 pKa = 11.03SGVGWNAFASVSGTGHH345 pKa = 6.44QIATEE350 pKa = 4.42TINGGSAYY358 pKa = 10.08AAALATQTGAARR370 pKa = 11.84NVLFSSEE377 pKa = 3.75GVMADD382 pKa = 4.33ANMLQQAISYY392 pKa = 8.11AANGTGLSVGLHH404 pKa = 6.58LSRR407 pKa = 11.84EE408 pKa = 4.38SGVVAARR415 pKa = 11.84VDD417 pKa = 3.45MDD419 pKa = 3.35QSQEE423 pKa = 4.17LEE425 pKa = 4.4DD426 pKa = 3.84VDD428 pKa = 5.0PPGSTPGIYY437 pKa = 10.21DD438 pKa = 3.48KK439 pKa = 11.03LLPILTQWKK448 pKa = 9.71ADD450 pKa = 3.66YY451 pKa = 10.88NFVGSFYY458 pKa = 11.03LNVGNGTNGTGTDD471 pKa = 3.38WAVSLPYY478 pKa = 10.58YY479 pKa = 10.32KK480 pKa = 10.85ALLDD484 pKa = 4.35LGNEE488 pKa = 4.38LGTHH492 pKa = 6.09SLTHH496 pKa = 7.0PDD498 pKa = 3.09NTNLLNAQQIQTEE511 pKa = 4.41FQDD514 pKa = 4.37SIAILNQQISAYY526 pKa = 10.08LNQPFQITGAAVPGAPEE543 pKa = 4.11SLPVSQEE550 pKa = 3.05IMKK553 pKa = 10.38YY554 pKa = 10.05VSDD557 pKa = 4.24YY558 pKa = 10.76LTGGYY563 pKa = 9.32AAQGAGYY570 pKa = 8.77PNAFGYY576 pKa = 7.41LTPADD581 pKa = 3.35QSKK584 pKa = 10.57IYY586 pKa = 9.4FAPNTSFDD594 pKa = 3.39YY595 pKa = 11.03TFIEE599 pKa = 4.92FQQKK603 pKa = 7.0TVAEE607 pKa = 4.48ATALWIGEE615 pKa = 3.92FDD617 pKa = 4.14KK618 pKa = 11.48LVANAGMPVIVWPWHH633 pKa = 6.94DD634 pKa = 3.57YY635 pKa = 10.36GAADD639 pKa = 4.58FLSGSPTVDD648 pKa = 2.81SPYY651 pKa = 10.57DD652 pKa = 3.35ISLFTSWIQHH662 pKa = 4.4AVAANMEE669 pKa = 4.58FVTLDD674 pKa = 3.39DD675 pKa = 3.79LAARR679 pKa = 11.84MTAVKK684 pKa = 10.21NATITTTVAGNSITASVGGANVGNLSLDD712 pKa = 3.64VEE714 pKa = 4.49RR715 pKa = 11.84QGALVIQNVTGWYY728 pKa = 9.97AYY730 pKa = 10.57DD731 pKa = 3.64SDD733 pKa = 5.55SVFLPQSGGTFKK745 pKa = 9.84ITLGAAADD753 pKa = 4.2DD754 pKa = 4.07VTHH757 pKa = 6.31ITDD760 pKa = 3.66LPMRR764 pKa = 11.84AVLLSVAGDD773 pKa = 3.42GHH775 pKa = 6.51NLAFSIQGEE784 pKa = 4.55GKK786 pKa = 10.33VVIDD790 pKa = 3.9IANPGTDD797 pKa = 3.75GIVVTGAAGYY807 pKa = 8.64SQVGEE812 pKa = 4.24IVTVDD817 pKa = 3.19LGGYY821 pKa = 8.58GLHH824 pKa = 6.52NVMLEE829 pKa = 4.09YY830 pKa = 10.67KK831 pKa = 9.21NPPVIVSNGGGPTATLSMGEE851 pKa = 4.19NGTAVTTVVATDD863 pKa = 3.59ADD865 pKa = 3.83VGTVIGYY872 pKa = 9.33SISGGADD879 pKa = 2.87AAKK882 pKa = 10.28FRR884 pKa = 11.84IDD886 pKa = 3.38GATGALTFLSAPNFEE901 pKa = 4.5AAGDD905 pKa = 3.74VGTDD909 pKa = 3.01NVYY912 pKa = 11.12DD913 pKa = 4.11VIVRR917 pKa = 11.84ASDD920 pKa = 3.12GTLYY924 pKa = 10.68DD925 pKa = 3.86DD926 pKa = 3.62QAIAVTITNVNEE938 pKa = 4.03APVITSNGGGASAAVSVAEE957 pKa = 4.16NLTGVTTVTAIDD969 pKa = 4.0PDD971 pKa = 4.25AGTMLAYY978 pKa = 10.45SIVGGPDD985 pKa = 2.74RR986 pKa = 11.84ALFSIDD992 pKa = 3.59PSTGALAFLSPPNYY1006 pKa = 10.02DD1007 pKa = 3.99LPGDD1011 pKa = 3.83ANGNNVYY1018 pKa = 10.62SLTVRR1023 pKa = 11.84VSDD1026 pKa = 3.91GALFDD1031 pKa = 4.18DD1032 pKa = 3.92QVINVTVTNVNEE1044 pKa = 3.96QAPVITSGGGGDD1056 pKa = 3.39TAAISVSSGTRR1067 pKa = 11.84TVMKK1071 pKa = 9.27VTAVDD1076 pKa = 3.51PDD1078 pKa = 4.0ANTTLAYY1085 pKa = 10.36SIAGGVDD1092 pKa = 2.76AGKK1095 pKa = 8.57FTINASTGQLNFVSTPNYY1113 pKa = 10.27LLPTDD1118 pKa = 4.13AGRR1121 pKa = 11.84DD1122 pKa = 3.39NVYY1125 pKa = 10.97DD1126 pKa = 4.04VIVRR1130 pKa = 11.84ASDD1133 pKa = 3.47GTLHH1137 pKa = 7.35DD1138 pKa = 4.0EE1139 pKa = 3.91QAIAVTVTILNAPPIIISNGGGASATIRR1167 pKa = 11.84MAEE1170 pKa = 4.19NGTAVTKK1177 pKa = 9.35VTATDD1182 pKa = 3.41VNLGQSITYY1191 pKa = 10.06SLGSAVDD1198 pKa = 3.7GALFSIDD1205 pKa = 3.73PQSGVLTFKK1214 pKa = 10.53SAPDD1218 pKa = 3.74FEE1220 pKa = 6.3APADD1224 pKa = 3.87SNLDD1228 pKa = 2.83NRR1230 pKa = 11.84YY1231 pKa = 8.85QVQVLATDD1239 pKa = 3.44SLGAQDD1245 pKa = 4.31FQNLTVVVNNVSGLSLTAASGGSTLIGQGEE1275 pKa = 4.67EE1276 pKa = 3.94DD1277 pKa = 3.82TLRR1280 pKa = 11.84GAGGNDD1286 pKa = 3.26TLIGNGGSDD1295 pKa = 3.73TLNGAGGNDD1304 pKa = 4.11NIDD1307 pKa = 3.63GGAGADD1313 pKa = 4.32RR1314 pKa = 11.84ITGGAGADD1322 pKa = 3.5ILTGGADD1329 pKa = 3.7ADD1331 pKa = 4.0LFIYY1335 pKa = 10.42NATGNSTLAVMDD1347 pKa = 5.38RR1348 pKa = 11.84IMDD1351 pKa = 4.11FQQGIDD1357 pKa = 3.95RR1358 pKa = 11.84IDD1360 pKa = 3.57LSGIDD1365 pKa = 4.14ALSALSGNQAFSFIGNAAFSGAGQLHH1391 pKa = 6.42VYY1393 pKa = 10.97SNGTDD1398 pKa = 3.18SFVEE1402 pKa = 4.04ANVDD1406 pKa = 3.7NNPATVDD1413 pKa = 3.7FAIRR1417 pKa = 11.84LIGVHH1422 pKa = 6.35NLTASDD1428 pKa = 3.96FLLL1431 pKa = 4.74
Molecular weight: 148.01 kDa
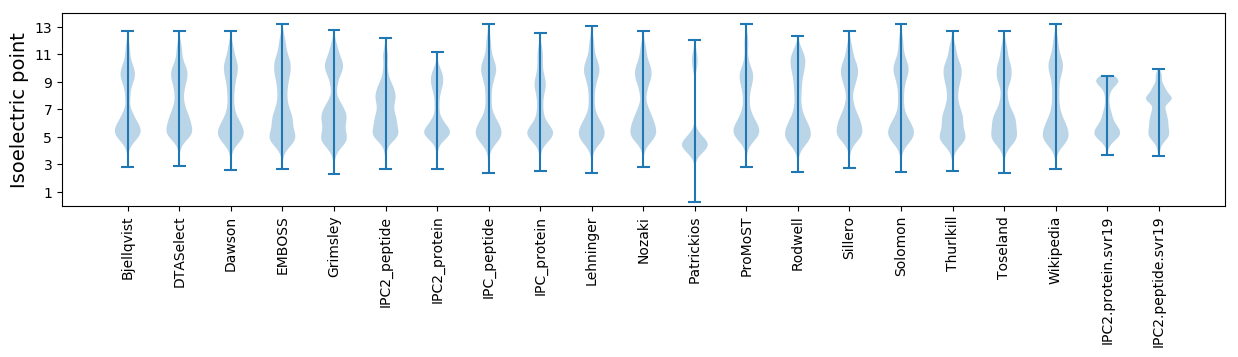
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D4YZ56|D4YZ56_SPHJU Uncharacterized protein OS=Sphingobium japonicum (strain DSM 16413 / CCM 7287 / MTCC 6362 / UT26 / NBRC 101211 / UT26S) OX=452662 GN=SJA_C1-08040 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84NVLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84NVLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1313854 |
26 |
4159 |
302.2 |
32.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.205 ± 0.061 | 0.811 ± 0.013 |
5.946 ± 0.037 | 5.353 ± 0.038 |
3.527 ± 0.024 | 9.0 ± 0.048 |
2.069 ± 0.021 | 5.045 ± 0.023 |
2.964 ± 0.028 | 9.98 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.557 ± 0.02 | 2.547 ± 0.03 |
5.347 ± 0.036 | 3.209 ± 0.023 |
7.664 ± 0.046 | 5.345 ± 0.029 |
4.9 ± 0.039 | 6.909 ± 0.029 |
1.421 ± 0.017 | 2.202 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
