
Beihai narna-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIC8|A0A1L3KIC8_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 2 OX=1922447 PE=4 SV=1
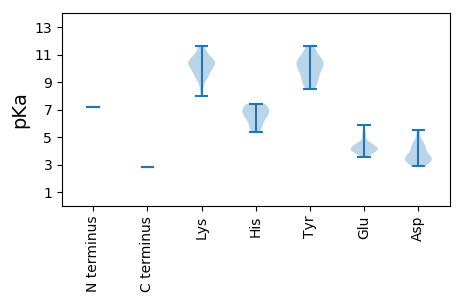
MM1 pKa = 7.15EE2 pKa = 4.13TTLAKK7 pKa = 10.56ALVEE11 pKa = 4.36SCVKK15 pKa = 10.61YY16 pKa = 9.99YY17 pKa = 10.66SPKK20 pKa = 9.48EE21 pKa = 3.55ASAYY25 pKa = 10.44LIEE28 pKa = 5.07FGVKK32 pKa = 9.56YY33 pKa = 10.33FGLSSCHH40 pKa = 7.35SPDD43 pKa = 3.51LWDD46 pKa = 3.6EE47 pKa = 4.1FLGRR51 pKa = 11.84FYY53 pKa = 10.47PSSDD57 pKa = 4.38SIPMDD62 pKa = 3.4GVTDD66 pKa = 3.5WRR68 pKa = 11.84SDD70 pKa = 3.36LSRR73 pKa = 11.84EE74 pKa = 4.04IVDD77 pKa = 3.44SYY79 pKa = 11.62EE80 pKa = 3.68KK81 pKa = 9.82VKK83 pKa = 10.94RR84 pKa = 11.84MFEE87 pKa = 4.1VYY89 pKa = 10.27SYY91 pKa = 10.88IINTRR96 pKa = 11.84LGQTLTWASQRR107 pKa = 11.84EE108 pKa = 4.41TMTRR112 pKa = 11.84WFRR115 pKa = 11.84YY116 pKa = 8.97ALRR119 pKa = 11.84GKK121 pKa = 9.92LEE123 pKa = 4.0KK124 pKa = 10.52VLKK127 pKa = 10.54FKK129 pKa = 10.02TATLLALGLNQSEE142 pKa = 4.46LPPRR146 pKa = 11.84PSCLEE151 pKa = 3.73ADD153 pKa = 3.77DD154 pKa = 4.06RR155 pKa = 11.84ANYY158 pKa = 9.57LFQGKK163 pKa = 8.39VGRR166 pKa = 11.84RR167 pKa = 11.84ILHH170 pKa = 5.39CTRR173 pKa = 11.84IVSAKK178 pKa = 9.28RR179 pKa = 11.84DD180 pKa = 3.43NGEE183 pKa = 3.83RR184 pKa = 11.84VQFLYY189 pKa = 10.75EE190 pKa = 4.25LYY192 pKa = 9.27SAKK195 pKa = 10.14RR196 pKa = 11.84ASLPVSSDD204 pKa = 3.18FVTEE208 pKa = 3.83ALEE211 pKa = 4.24KK212 pKa = 10.46HH213 pKa = 5.53WFTLTEE219 pKa = 4.65LDD221 pKa = 5.51QKK223 pKa = 11.61DD224 pKa = 4.15LDD226 pKa = 4.36NPFLDD231 pKa = 4.66DD232 pKa = 5.23LIEE235 pKa = 4.33EE236 pKa = 4.44VTRR239 pKa = 11.84TVDD242 pKa = 3.44EE243 pKa = 5.3LIPPVKK249 pKa = 9.63CTPHH253 pKa = 6.42YY254 pKa = 9.92KK255 pKa = 9.59VCPSFGASFKK265 pKa = 10.16TGRR268 pKa = 11.84DD269 pKa = 2.91KK270 pKa = 11.52GGSYY274 pKa = 10.62SEE276 pKa = 4.73LVGEE280 pKa = 4.51SQLTSNEE287 pKa = 3.53GHH289 pKa = 7.15LGGFMQKK296 pKa = 9.56SCPILVQEE304 pKa = 4.24SFDD307 pKa = 3.52WKK309 pKa = 11.16SEE311 pKa = 3.94ITSKK315 pKa = 10.3PSKK318 pKa = 10.34IGGGDD323 pKa = 3.33VMGVEE328 pKa = 4.15WRR330 pKa = 11.84VPDD333 pKa = 4.78VDD335 pKa = 5.24EE336 pKa = 5.23DD337 pKa = 3.47WLDD340 pKa = 3.82YY341 pKa = 11.47VSVASQLQDD350 pKa = 2.86IDD352 pKa = 4.08CRR354 pKa = 11.84PVALLEE360 pKa = 4.09PFKK363 pKa = 11.27VRR365 pKa = 11.84VITRR369 pKa = 11.84GSAPAYY375 pKa = 9.3QLARR379 pKa = 11.84AYY381 pKa = 10.39QRR383 pKa = 11.84IIAPLVGKK391 pKa = 10.16NPAFRR396 pKa = 11.84LTRR399 pKa = 11.84GPAKK403 pKa = 9.4MDD405 pKa = 4.56HH406 pKa = 6.75INDD409 pKa = 4.27LLEE412 pKa = 4.38KK413 pKa = 10.27AQGPSAEE420 pKa = 4.18YY421 pKa = 10.34IEE423 pKa = 5.5AFEE426 pKa = 4.45CGARR430 pKa = 11.84RR431 pKa = 11.84STLEE435 pKa = 3.94AEE437 pKa = 5.07DD438 pKa = 4.39YY439 pKa = 11.44SSGTWRR445 pKa = 11.84PEE447 pKa = 4.3GIWVSGDD454 pKa = 3.41YY455 pKa = 10.96EE456 pKa = 4.57SATDD460 pKa = 4.49LLNATLSEE468 pKa = 4.05ICLDD472 pKa = 4.5RR473 pKa = 11.84ILHH476 pKa = 6.01NLDD479 pKa = 2.92APYY482 pKa = 10.64EE483 pKa = 4.06HH484 pKa = 7.26RR485 pKa = 11.84RR486 pKa = 11.84TLFGCLTRR494 pKa = 11.84HH495 pKa = 5.65TLHH498 pKa = 6.77SADD501 pKa = 4.21DD502 pKa = 4.13SKK504 pKa = 11.58VAPQINGQLMGSPLSFPILCMVNAAVTRR532 pKa = 11.84KK533 pKa = 10.09AIEE536 pKa = 3.97VCTGRR541 pKa = 11.84KK542 pKa = 9.19KK543 pKa = 10.62LLKK546 pKa = 10.32DD547 pKa = 3.1HH548 pKa = 7.0AMLINGDD555 pKa = 3.72DD556 pKa = 3.67VLFHH560 pKa = 6.53LRR562 pKa = 11.84SIAEE566 pKa = 4.02YY567 pKa = 9.79EE568 pKa = 4.01VWKK571 pKa = 9.22TFTAGAGLKK580 pKa = 10.42FSLGKK585 pKa = 10.39NYY587 pKa = 9.94VSRR590 pKa = 11.84SYY592 pKa = 11.22LVINSEE598 pKa = 3.82IFKK601 pKa = 10.35IRR603 pKa = 11.84PHH605 pKa = 6.94VNFFGEE611 pKa = 4.49RR612 pKa = 11.84KK613 pKa = 8.82WICDD617 pKa = 3.05RR618 pKa = 11.84SLPIINMGHH627 pKa = 6.24VYY629 pKa = 9.36VQKK632 pKa = 10.65KK633 pKa = 10.24SSSKK637 pKa = 10.11LQSEE641 pKa = 3.93QSMYY645 pKa = 8.64GTHH648 pKa = 6.46YY649 pKa = 10.02LQKK652 pKa = 10.99DD653 pKa = 3.76SLKK656 pKa = 10.78DD657 pKa = 3.32SCEE660 pKa = 4.35DD661 pKa = 3.24FVEE664 pKa = 4.07AHH666 pKa = 6.86PLDD669 pKa = 3.93KK670 pKa = 10.92RR671 pKa = 11.84CFALSKK677 pKa = 9.55WISARR682 pKa = 11.84KK683 pKa = 9.55HH684 pKa = 5.97EE685 pKa = 5.87LDD687 pKa = 3.08TCLPKK692 pKa = 10.67GMSYY696 pKa = 10.32FVHH699 pKa = 6.83EE700 pKa = 4.51SLGGLGLPVYY710 pKa = 8.6PHH712 pKa = 7.41KK713 pKa = 10.88DD714 pKa = 3.14QLVGITEE721 pKa = 4.41GQRR724 pKa = 11.84KK725 pKa = 7.99LAAYY729 pKa = 9.33LWTSQDD735 pKa = 2.89PTARR739 pKa = 11.84LINTGALRR747 pKa = 11.84LPIPSFVEE755 pKa = 4.1RR756 pKa = 11.84YY757 pKa = 8.85MRR759 pKa = 11.84DD760 pKa = 3.02KK761 pKa = 11.41SDD763 pKa = 3.15LLQRR767 pKa = 11.84LDD769 pKa = 4.72VRR771 pKa = 11.84YY772 pKa = 8.49EE773 pKa = 3.63WSNVKK778 pKa = 10.1VKK780 pKa = 10.63EE781 pKa = 4.08DD782 pKa = 3.17WPDD785 pKa = 3.42FHH787 pKa = 7.41RR788 pKa = 11.84WTAVGIVGQDD798 pKa = 3.55PPDD801 pKa = 3.46MKK803 pKa = 10.39PFRR806 pKa = 11.84RR807 pKa = 11.84VYY809 pKa = 9.03EE810 pKa = 4.1AVWRR814 pKa = 11.84DD815 pKa = 3.62AQASWASPLSFRR827 pKa = 11.84KK828 pKa = 9.81CIKK831 pKa = 10.45GVFDD835 pKa = 3.96GVPGYY840 pKa = 9.98FGPAEE845 pKa = 3.98RR846 pKa = 11.84VRR848 pKa = 11.84KK849 pKa = 8.85QQ850 pKa = 2.84
MM1 pKa = 7.15EE2 pKa = 4.13TTLAKK7 pKa = 10.56ALVEE11 pKa = 4.36SCVKK15 pKa = 10.61YY16 pKa = 9.99YY17 pKa = 10.66SPKK20 pKa = 9.48EE21 pKa = 3.55ASAYY25 pKa = 10.44LIEE28 pKa = 5.07FGVKK32 pKa = 9.56YY33 pKa = 10.33FGLSSCHH40 pKa = 7.35SPDD43 pKa = 3.51LWDD46 pKa = 3.6EE47 pKa = 4.1FLGRR51 pKa = 11.84FYY53 pKa = 10.47PSSDD57 pKa = 4.38SIPMDD62 pKa = 3.4GVTDD66 pKa = 3.5WRR68 pKa = 11.84SDD70 pKa = 3.36LSRR73 pKa = 11.84EE74 pKa = 4.04IVDD77 pKa = 3.44SYY79 pKa = 11.62EE80 pKa = 3.68KK81 pKa = 9.82VKK83 pKa = 10.94RR84 pKa = 11.84MFEE87 pKa = 4.1VYY89 pKa = 10.27SYY91 pKa = 10.88IINTRR96 pKa = 11.84LGQTLTWASQRR107 pKa = 11.84EE108 pKa = 4.41TMTRR112 pKa = 11.84WFRR115 pKa = 11.84YY116 pKa = 8.97ALRR119 pKa = 11.84GKK121 pKa = 9.92LEE123 pKa = 4.0KK124 pKa = 10.52VLKK127 pKa = 10.54FKK129 pKa = 10.02TATLLALGLNQSEE142 pKa = 4.46LPPRR146 pKa = 11.84PSCLEE151 pKa = 3.73ADD153 pKa = 3.77DD154 pKa = 4.06RR155 pKa = 11.84ANYY158 pKa = 9.57LFQGKK163 pKa = 8.39VGRR166 pKa = 11.84RR167 pKa = 11.84ILHH170 pKa = 5.39CTRR173 pKa = 11.84IVSAKK178 pKa = 9.28RR179 pKa = 11.84DD180 pKa = 3.43NGEE183 pKa = 3.83RR184 pKa = 11.84VQFLYY189 pKa = 10.75EE190 pKa = 4.25LYY192 pKa = 9.27SAKK195 pKa = 10.14RR196 pKa = 11.84ASLPVSSDD204 pKa = 3.18FVTEE208 pKa = 3.83ALEE211 pKa = 4.24KK212 pKa = 10.46HH213 pKa = 5.53WFTLTEE219 pKa = 4.65LDD221 pKa = 5.51QKK223 pKa = 11.61DD224 pKa = 4.15LDD226 pKa = 4.36NPFLDD231 pKa = 4.66DD232 pKa = 5.23LIEE235 pKa = 4.33EE236 pKa = 4.44VTRR239 pKa = 11.84TVDD242 pKa = 3.44EE243 pKa = 5.3LIPPVKK249 pKa = 9.63CTPHH253 pKa = 6.42YY254 pKa = 9.92KK255 pKa = 9.59VCPSFGASFKK265 pKa = 10.16TGRR268 pKa = 11.84DD269 pKa = 2.91KK270 pKa = 11.52GGSYY274 pKa = 10.62SEE276 pKa = 4.73LVGEE280 pKa = 4.51SQLTSNEE287 pKa = 3.53GHH289 pKa = 7.15LGGFMQKK296 pKa = 9.56SCPILVQEE304 pKa = 4.24SFDD307 pKa = 3.52WKK309 pKa = 11.16SEE311 pKa = 3.94ITSKK315 pKa = 10.3PSKK318 pKa = 10.34IGGGDD323 pKa = 3.33VMGVEE328 pKa = 4.15WRR330 pKa = 11.84VPDD333 pKa = 4.78VDD335 pKa = 5.24EE336 pKa = 5.23DD337 pKa = 3.47WLDD340 pKa = 3.82YY341 pKa = 11.47VSVASQLQDD350 pKa = 2.86IDD352 pKa = 4.08CRR354 pKa = 11.84PVALLEE360 pKa = 4.09PFKK363 pKa = 11.27VRR365 pKa = 11.84VITRR369 pKa = 11.84GSAPAYY375 pKa = 9.3QLARR379 pKa = 11.84AYY381 pKa = 10.39QRR383 pKa = 11.84IIAPLVGKK391 pKa = 10.16NPAFRR396 pKa = 11.84LTRR399 pKa = 11.84GPAKK403 pKa = 9.4MDD405 pKa = 4.56HH406 pKa = 6.75INDD409 pKa = 4.27LLEE412 pKa = 4.38KK413 pKa = 10.27AQGPSAEE420 pKa = 4.18YY421 pKa = 10.34IEE423 pKa = 5.5AFEE426 pKa = 4.45CGARR430 pKa = 11.84RR431 pKa = 11.84STLEE435 pKa = 3.94AEE437 pKa = 5.07DD438 pKa = 4.39YY439 pKa = 11.44SSGTWRR445 pKa = 11.84PEE447 pKa = 4.3GIWVSGDD454 pKa = 3.41YY455 pKa = 10.96EE456 pKa = 4.57SATDD460 pKa = 4.49LLNATLSEE468 pKa = 4.05ICLDD472 pKa = 4.5RR473 pKa = 11.84ILHH476 pKa = 6.01NLDD479 pKa = 2.92APYY482 pKa = 10.64EE483 pKa = 4.06HH484 pKa = 7.26RR485 pKa = 11.84RR486 pKa = 11.84TLFGCLTRR494 pKa = 11.84HH495 pKa = 5.65TLHH498 pKa = 6.77SADD501 pKa = 4.21DD502 pKa = 4.13SKK504 pKa = 11.58VAPQINGQLMGSPLSFPILCMVNAAVTRR532 pKa = 11.84KK533 pKa = 10.09AIEE536 pKa = 3.97VCTGRR541 pKa = 11.84KK542 pKa = 9.19KK543 pKa = 10.62LLKK546 pKa = 10.32DD547 pKa = 3.1HH548 pKa = 7.0AMLINGDD555 pKa = 3.72DD556 pKa = 3.67VLFHH560 pKa = 6.53LRR562 pKa = 11.84SIAEE566 pKa = 4.02YY567 pKa = 9.79EE568 pKa = 4.01VWKK571 pKa = 9.22TFTAGAGLKK580 pKa = 10.42FSLGKK585 pKa = 10.39NYY587 pKa = 9.94VSRR590 pKa = 11.84SYY592 pKa = 11.22LVINSEE598 pKa = 3.82IFKK601 pKa = 10.35IRR603 pKa = 11.84PHH605 pKa = 6.94VNFFGEE611 pKa = 4.49RR612 pKa = 11.84KK613 pKa = 8.82WICDD617 pKa = 3.05RR618 pKa = 11.84SLPIINMGHH627 pKa = 6.24VYY629 pKa = 9.36VQKK632 pKa = 10.65KK633 pKa = 10.24SSSKK637 pKa = 10.11LQSEE641 pKa = 3.93QSMYY645 pKa = 8.64GTHH648 pKa = 6.46YY649 pKa = 10.02LQKK652 pKa = 10.99DD653 pKa = 3.76SLKK656 pKa = 10.78DD657 pKa = 3.32SCEE660 pKa = 4.35DD661 pKa = 3.24FVEE664 pKa = 4.07AHH666 pKa = 6.86PLDD669 pKa = 3.93KK670 pKa = 10.92RR671 pKa = 11.84CFALSKK677 pKa = 9.55WISARR682 pKa = 11.84KK683 pKa = 9.55HH684 pKa = 5.97EE685 pKa = 5.87LDD687 pKa = 3.08TCLPKK692 pKa = 10.67GMSYY696 pKa = 10.32FVHH699 pKa = 6.83EE700 pKa = 4.51SLGGLGLPVYY710 pKa = 8.6PHH712 pKa = 7.41KK713 pKa = 10.88DD714 pKa = 3.14QLVGITEE721 pKa = 4.41GQRR724 pKa = 11.84KK725 pKa = 7.99LAAYY729 pKa = 9.33LWTSQDD735 pKa = 2.89PTARR739 pKa = 11.84LINTGALRR747 pKa = 11.84LPIPSFVEE755 pKa = 4.1RR756 pKa = 11.84YY757 pKa = 8.85MRR759 pKa = 11.84DD760 pKa = 3.02KK761 pKa = 11.41SDD763 pKa = 3.15LLQRR767 pKa = 11.84LDD769 pKa = 4.72VRR771 pKa = 11.84YY772 pKa = 8.49EE773 pKa = 3.63WSNVKK778 pKa = 10.1VKK780 pKa = 10.63EE781 pKa = 4.08DD782 pKa = 3.17WPDD785 pKa = 3.42FHH787 pKa = 7.41RR788 pKa = 11.84WTAVGIVGQDD798 pKa = 3.55PPDD801 pKa = 3.46MKK803 pKa = 10.39PFRR806 pKa = 11.84RR807 pKa = 11.84VYY809 pKa = 9.03EE810 pKa = 4.1AVWRR814 pKa = 11.84DD815 pKa = 3.62AQASWASPLSFRR827 pKa = 11.84KK828 pKa = 9.81CIKK831 pKa = 10.45GVFDD835 pKa = 3.96GVPGYY840 pKa = 9.98FGPAEE845 pKa = 3.98RR846 pKa = 11.84VRR848 pKa = 11.84KK849 pKa = 8.85QQ850 pKa = 2.84
Molecular weight: 96.65 kDa
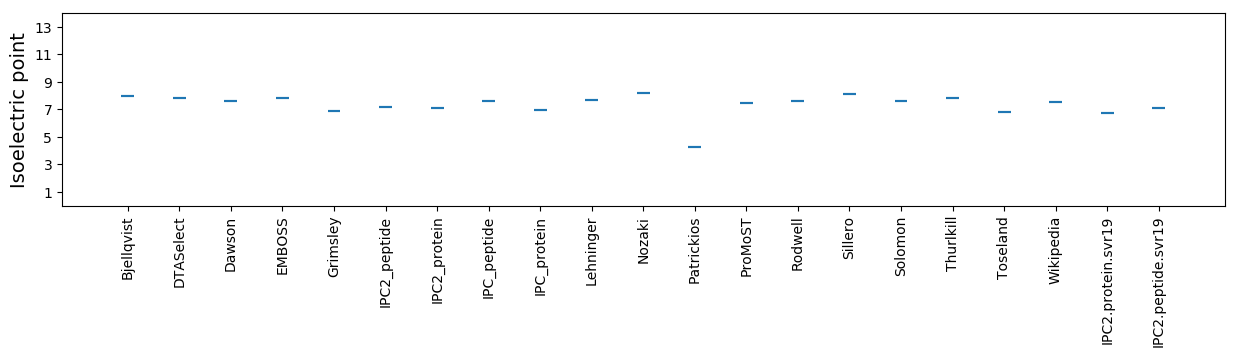
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIC8|A0A1L3KIC8_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 2 OX=1922447 PE=4 SV=1
MM1 pKa = 7.15EE2 pKa = 4.13TTLAKK7 pKa = 10.56ALVEE11 pKa = 4.36SCVKK15 pKa = 10.61YY16 pKa = 9.99YY17 pKa = 10.66SPKK20 pKa = 9.48EE21 pKa = 3.55ASAYY25 pKa = 10.44LIEE28 pKa = 5.07FGVKK32 pKa = 9.56YY33 pKa = 10.33FGLSSCHH40 pKa = 7.35SPDD43 pKa = 3.51LWDD46 pKa = 3.6EE47 pKa = 4.1FLGRR51 pKa = 11.84FYY53 pKa = 10.47PSSDD57 pKa = 4.38SIPMDD62 pKa = 3.4GVTDD66 pKa = 3.5WRR68 pKa = 11.84SDD70 pKa = 3.36LSRR73 pKa = 11.84EE74 pKa = 4.04IVDD77 pKa = 3.44SYY79 pKa = 11.62EE80 pKa = 3.68KK81 pKa = 9.82VKK83 pKa = 10.94RR84 pKa = 11.84MFEE87 pKa = 4.1VYY89 pKa = 10.27SYY91 pKa = 10.88IINTRR96 pKa = 11.84LGQTLTWASQRR107 pKa = 11.84EE108 pKa = 4.41TMTRR112 pKa = 11.84WFRR115 pKa = 11.84YY116 pKa = 8.97ALRR119 pKa = 11.84GKK121 pKa = 9.92LEE123 pKa = 4.0KK124 pKa = 10.52VLKK127 pKa = 10.54FKK129 pKa = 10.02TATLLALGLNQSEE142 pKa = 4.46LPPRR146 pKa = 11.84PSCLEE151 pKa = 3.73ADD153 pKa = 3.77DD154 pKa = 4.06RR155 pKa = 11.84ANYY158 pKa = 9.57LFQGKK163 pKa = 8.39VGRR166 pKa = 11.84RR167 pKa = 11.84ILHH170 pKa = 5.39CTRR173 pKa = 11.84IVSAKK178 pKa = 9.28RR179 pKa = 11.84DD180 pKa = 3.43NGEE183 pKa = 3.83RR184 pKa = 11.84VQFLYY189 pKa = 10.75EE190 pKa = 4.25LYY192 pKa = 9.27SAKK195 pKa = 10.14RR196 pKa = 11.84ASLPVSSDD204 pKa = 3.18FVTEE208 pKa = 3.83ALEE211 pKa = 4.24KK212 pKa = 10.46HH213 pKa = 5.53WFTLTEE219 pKa = 4.65LDD221 pKa = 5.51QKK223 pKa = 11.61DD224 pKa = 4.15LDD226 pKa = 4.36NPFLDD231 pKa = 4.66DD232 pKa = 5.23LIEE235 pKa = 4.33EE236 pKa = 4.44VTRR239 pKa = 11.84TVDD242 pKa = 3.44EE243 pKa = 5.3LIPPVKK249 pKa = 9.63CTPHH253 pKa = 6.42YY254 pKa = 9.92KK255 pKa = 9.59VCPSFGASFKK265 pKa = 10.16TGRR268 pKa = 11.84DD269 pKa = 2.91KK270 pKa = 11.52GGSYY274 pKa = 10.62SEE276 pKa = 4.73LVGEE280 pKa = 4.51SQLTSNEE287 pKa = 3.53GHH289 pKa = 7.15LGGFMQKK296 pKa = 9.56SCPILVQEE304 pKa = 4.24SFDD307 pKa = 3.52WKK309 pKa = 11.16SEE311 pKa = 3.94ITSKK315 pKa = 10.3PSKK318 pKa = 10.34IGGGDD323 pKa = 3.33VMGVEE328 pKa = 4.15WRR330 pKa = 11.84VPDD333 pKa = 4.78VDD335 pKa = 5.24EE336 pKa = 5.23DD337 pKa = 3.47WLDD340 pKa = 3.82YY341 pKa = 11.47VSVASQLQDD350 pKa = 2.86IDD352 pKa = 4.08CRR354 pKa = 11.84PVALLEE360 pKa = 4.09PFKK363 pKa = 11.27VRR365 pKa = 11.84VITRR369 pKa = 11.84GSAPAYY375 pKa = 9.3QLARR379 pKa = 11.84AYY381 pKa = 10.39QRR383 pKa = 11.84IIAPLVGKK391 pKa = 10.16NPAFRR396 pKa = 11.84LTRR399 pKa = 11.84GPAKK403 pKa = 9.4MDD405 pKa = 4.56HH406 pKa = 6.75INDD409 pKa = 4.27LLEE412 pKa = 4.38KK413 pKa = 10.27AQGPSAEE420 pKa = 4.18YY421 pKa = 10.34IEE423 pKa = 5.5AFEE426 pKa = 4.45CGARR430 pKa = 11.84RR431 pKa = 11.84STLEE435 pKa = 3.94AEE437 pKa = 5.07DD438 pKa = 4.39YY439 pKa = 11.44SSGTWRR445 pKa = 11.84PEE447 pKa = 4.3GIWVSGDD454 pKa = 3.41YY455 pKa = 10.96EE456 pKa = 4.57SATDD460 pKa = 4.49LLNATLSEE468 pKa = 4.05ICLDD472 pKa = 4.5RR473 pKa = 11.84ILHH476 pKa = 6.01NLDD479 pKa = 2.92APYY482 pKa = 10.64EE483 pKa = 4.06HH484 pKa = 7.26RR485 pKa = 11.84RR486 pKa = 11.84TLFGCLTRR494 pKa = 11.84HH495 pKa = 5.65TLHH498 pKa = 6.77SADD501 pKa = 4.21DD502 pKa = 4.13SKK504 pKa = 11.58VAPQINGQLMGSPLSFPILCMVNAAVTRR532 pKa = 11.84KK533 pKa = 10.09AIEE536 pKa = 3.97VCTGRR541 pKa = 11.84KK542 pKa = 9.19KK543 pKa = 10.62LLKK546 pKa = 10.32DD547 pKa = 3.1HH548 pKa = 7.0AMLINGDD555 pKa = 3.72DD556 pKa = 3.67VLFHH560 pKa = 6.53LRR562 pKa = 11.84SIAEE566 pKa = 4.02YY567 pKa = 9.79EE568 pKa = 4.01VWKK571 pKa = 9.22TFTAGAGLKK580 pKa = 10.42FSLGKK585 pKa = 10.39NYY587 pKa = 9.94VSRR590 pKa = 11.84SYY592 pKa = 11.22LVINSEE598 pKa = 3.82IFKK601 pKa = 10.35IRR603 pKa = 11.84PHH605 pKa = 6.94VNFFGEE611 pKa = 4.49RR612 pKa = 11.84KK613 pKa = 8.82WICDD617 pKa = 3.05RR618 pKa = 11.84SLPIINMGHH627 pKa = 6.24VYY629 pKa = 9.36VQKK632 pKa = 10.65KK633 pKa = 10.24SSSKK637 pKa = 10.11LQSEE641 pKa = 3.93QSMYY645 pKa = 8.64GTHH648 pKa = 6.46YY649 pKa = 10.02LQKK652 pKa = 10.99DD653 pKa = 3.76SLKK656 pKa = 10.78DD657 pKa = 3.32SCEE660 pKa = 4.35DD661 pKa = 3.24FVEE664 pKa = 4.07AHH666 pKa = 6.86PLDD669 pKa = 3.93KK670 pKa = 10.92RR671 pKa = 11.84CFALSKK677 pKa = 9.55WISARR682 pKa = 11.84KK683 pKa = 9.55HH684 pKa = 5.97EE685 pKa = 5.87LDD687 pKa = 3.08TCLPKK692 pKa = 10.67GMSYY696 pKa = 10.32FVHH699 pKa = 6.83EE700 pKa = 4.51SLGGLGLPVYY710 pKa = 8.6PHH712 pKa = 7.41KK713 pKa = 10.88DD714 pKa = 3.14QLVGITEE721 pKa = 4.41GQRR724 pKa = 11.84KK725 pKa = 7.99LAAYY729 pKa = 9.33LWTSQDD735 pKa = 2.89PTARR739 pKa = 11.84LINTGALRR747 pKa = 11.84LPIPSFVEE755 pKa = 4.1RR756 pKa = 11.84YY757 pKa = 8.85MRR759 pKa = 11.84DD760 pKa = 3.02KK761 pKa = 11.41SDD763 pKa = 3.15LLQRR767 pKa = 11.84LDD769 pKa = 4.72VRR771 pKa = 11.84YY772 pKa = 8.49EE773 pKa = 3.63WSNVKK778 pKa = 10.1VKK780 pKa = 10.63EE781 pKa = 4.08DD782 pKa = 3.17WPDD785 pKa = 3.42FHH787 pKa = 7.41RR788 pKa = 11.84WTAVGIVGQDD798 pKa = 3.55PPDD801 pKa = 3.46MKK803 pKa = 10.39PFRR806 pKa = 11.84RR807 pKa = 11.84VYY809 pKa = 9.03EE810 pKa = 4.1AVWRR814 pKa = 11.84DD815 pKa = 3.62AQASWASPLSFRR827 pKa = 11.84KK828 pKa = 9.81CIKK831 pKa = 10.45GVFDD835 pKa = 3.96GVPGYY840 pKa = 9.98FGPAEE845 pKa = 3.98RR846 pKa = 11.84VRR848 pKa = 11.84KK849 pKa = 8.85QQ850 pKa = 2.84
MM1 pKa = 7.15EE2 pKa = 4.13TTLAKK7 pKa = 10.56ALVEE11 pKa = 4.36SCVKK15 pKa = 10.61YY16 pKa = 9.99YY17 pKa = 10.66SPKK20 pKa = 9.48EE21 pKa = 3.55ASAYY25 pKa = 10.44LIEE28 pKa = 5.07FGVKK32 pKa = 9.56YY33 pKa = 10.33FGLSSCHH40 pKa = 7.35SPDD43 pKa = 3.51LWDD46 pKa = 3.6EE47 pKa = 4.1FLGRR51 pKa = 11.84FYY53 pKa = 10.47PSSDD57 pKa = 4.38SIPMDD62 pKa = 3.4GVTDD66 pKa = 3.5WRR68 pKa = 11.84SDD70 pKa = 3.36LSRR73 pKa = 11.84EE74 pKa = 4.04IVDD77 pKa = 3.44SYY79 pKa = 11.62EE80 pKa = 3.68KK81 pKa = 9.82VKK83 pKa = 10.94RR84 pKa = 11.84MFEE87 pKa = 4.1VYY89 pKa = 10.27SYY91 pKa = 10.88IINTRR96 pKa = 11.84LGQTLTWASQRR107 pKa = 11.84EE108 pKa = 4.41TMTRR112 pKa = 11.84WFRR115 pKa = 11.84YY116 pKa = 8.97ALRR119 pKa = 11.84GKK121 pKa = 9.92LEE123 pKa = 4.0KK124 pKa = 10.52VLKK127 pKa = 10.54FKK129 pKa = 10.02TATLLALGLNQSEE142 pKa = 4.46LPPRR146 pKa = 11.84PSCLEE151 pKa = 3.73ADD153 pKa = 3.77DD154 pKa = 4.06RR155 pKa = 11.84ANYY158 pKa = 9.57LFQGKK163 pKa = 8.39VGRR166 pKa = 11.84RR167 pKa = 11.84ILHH170 pKa = 5.39CTRR173 pKa = 11.84IVSAKK178 pKa = 9.28RR179 pKa = 11.84DD180 pKa = 3.43NGEE183 pKa = 3.83RR184 pKa = 11.84VQFLYY189 pKa = 10.75EE190 pKa = 4.25LYY192 pKa = 9.27SAKK195 pKa = 10.14RR196 pKa = 11.84ASLPVSSDD204 pKa = 3.18FVTEE208 pKa = 3.83ALEE211 pKa = 4.24KK212 pKa = 10.46HH213 pKa = 5.53WFTLTEE219 pKa = 4.65LDD221 pKa = 5.51QKK223 pKa = 11.61DD224 pKa = 4.15LDD226 pKa = 4.36NPFLDD231 pKa = 4.66DD232 pKa = 5.23LIEE235 pKa = 4.33EE236 pKa = 4.44VTRR239 pKa = 11.84TVDD242 pKa = 3.44EE243 pKa = 5.3LIPPVKK249 pKa = 9.63CTPHH253 pKa = 6.42YY254 pKa = 9.92KK255 pKa = 9.59VCPSFGASFKK265 pKa = 10.16TGRR268 pKa = 11.84DD269 pKa = 2.91KK270 pKa = 11.52GGSYY274 pKa = 10.62SEE276 pKa = 4.73LVGEE280 pKa = 4.51SQLTSNEE287 pKa = 3.53GHH289 pKa = 7.15LGGFMQKK296 pKa = 9.56SCPILVQEE304 pKa = 4.24SFDD307 pKa = 3.52WKK309 pKa = 11.16SEE311 pKa = 3.94ITSKK315 pKa = 10.3PSKK318 pKa = 10.34IGGGDD323 pKa = 3.33VMGVEE328 pKa = 4.15WRR330 pKa = 11.84VPDD333 pKa = 4.78VDD335 pKa = 5.24EE336 pKa = 5.23DD337 pKa = 3.47WLDD340 pKa = 3.82YY341 pKa = 11.47VSVASQLQDD350 pKa = 2.86IDD352 pKa = 4.08CRR354 pKa = 11.84PVALLEE360 pKa = 4.09PFKK363 pKa = 11.27VRR365 pKa = 11.84VITRR369 pKa = 11.84GSAPAYY375 pKa = 9.3QLARR379 pKa = 11.84AYY381 pKa = 10.39QRR383 pKa = 11.84IIAPLVGKK391 pKa = 10.16NPAFRR396 pKa = 11.84LTRR399 pKa = 11.84GPAKK403 pKa = 9.4MDD405 pKa = 4.56HH406 pKa = 6.75INDD409 pKa = 4.27LLEE412 pKa = 4.38KK413 pKa = 10.27AQGPSAEE420 pKa = 4.18YY421 pKa = 10.34IEE423 pKa = 5.5AFEE426 pKa = 4.45CGARR430 pKa = 11.84RR431 pKa = 11.84STLEE435 pKa = 3.94AEE437 pKa = 5.07DD438 pKa = 4.39YY439 pKa = 11.44SSGTWRR445 pKa = 11.84PEE447 pKa = 4.3GIWVSGDD454 pKa = 3.41YY455 pKa = 10.96EE456 pKa = 4.57SATDD460 pKa = 4.49LLNATLSEE468 pKa = 4.05ICLDD472 pKa = 4.5RR473 pKa = 11.84ILHH476 pKa = 6.01NLDD479 pKa = 2.92APYY482 pKa = 10.64EE483 pKa = 4.06HH484 pKa = 7.26RR485 pKa = 11.84RR486 pKa = 11.84TLFGCLTRR494 pKa = 11.84HH495 pKa = 5.65TLHH498 pKa = 6.77SADD501 pKa = 4.21DD502 pKa = 4.13SKK504 pKa = 11.58VAPQINGQLMGSPLSFPILCMVNAAVTRR532 pKa = 11.84KK533 pKa = 10.09AIEE536 pKa = 3.97VCTGRR541 pKa = 11.84KK542 pKa = 9.19KK543 pKa = 10.62LLKK546 pKa = 10.32DD547 pKa = 3.1HH548 pKa = 7.0AMLINGDD555 pKa = 3.72DD556 pKa = 3.67VLFHH560 pKa = 6.53LRR562 pKa = 11.84SIAEE566 pKa = 4.02YY567 pKa = 9.79EE568 pKa = 4.01VWKK571 pKa = 9.22TFTAGAGLKK580 pKa = 10.42FSLGKK585 pKa = 10.39NYY587 pKa = 9.94VSRR590 pKa = 11.84SYY592 pKa = 11.22LVINSEE598 pKa = 3.82IFKK601 pKa = 10.35IRR603 pKa = 11.84PHH605 pKa = 6.94VNFFGEE611 pKa = 4.49RR612 pKa = 11.84KK613 pKa = 8.82WICDD617 pKa = 3.05RR618 pKa = 11.84SLPIINMGHH627 pKa = 6.24VYY629 pKa = 9.36VQKK632 pKa = 10.65KK633 pKa = 10.24SSSKK637 pKa = 10.11LQSEE641 pKa = 3.93QSMYY645 pKa = 8.64GTHH648 pKa = 6.46YY649 pKa = 10.02LQKK652 pKa = 10.99DD653 pKa = 3.76SLKK656 pKa = 10.78DD657 pKa = 3.32SCEE660 pKa = 4.35DD661 pKa = 3.24FVEE664 pKa = 4.07AHH666 pKa = 6.86PLDD669 pKa = 3.93KK670 pKa = 10.92RR671 pKa = 11.84CFALSKK677 pKa = 9.55WISARR682 pKa = 11.84KK683 pKa = 9.55HH684 pKa = 5.97EE685 pKa = 5.87LDD687 pKa = 3.08TCLPKK692 pKa = 10.67GMSYY696 pKa = 10.32FVHH699 pKa = 6.83EE700 pKa = 4.51SLGGLGLPVYY710 pKa = 8.6PHH712 pKa = 7.41KK713 pKa = 10.88DD714 pKa = 3.14QLVGITEE721 pKa = 4.41GQRR724 pKa = 11.84KK725 pKa = 7.99LAAYY729 pKa = 9.33LWTSQDD735 pKa = 2.89PTARR739 pKa = 11.84LINTGALRR747 pKa = 11.84LPIPSFVEE755 pKa = 4.1RR756 pKa = 11.84YY757 pKa = 8.85MRR759 pKa = 11.84DD760 pKa = 3.02KK761 pKa = 11.41SDD763 pKa = 3.15LLQRR767 pKa = 11.84LDD769 pKa = 4.72VRR771 pKa = 11.84YY772 pKa = 8.49EE773 pKa = 3.63WSNVKK778 pKa = 10.1VKK780 pKa = 10.63EE781 pKa = 4.08DD782 pKa = 3.17WPDD785 pKa = 3.42FHH787 pKa = 7.41RR788 pKa = 11.84WTAVGIVGQDD798 pKa = 3.55PPDD801 pKa = 3.46MKK803 pKa = 10.39PFRR806 pKa = 11.84RR807 pKa = 11.84VYY809 pKa = 9.03EE810 pKa = 4.1AVWRR814 pKa = 11.84DD815 pKa = 3.62AQASWASPLSFRR827 pKa = 11.84KK828 pKa = 9.81CIKK831 pKa = 10.45GVFDD835 pKa = 3.96GVPGYY840 pKa = 9.98FGPAEE845 pKa = 3.98RR846 pKa = 11.84VRR848 pKa = 11.84KK849 pKa = 8.85QQ850 pKa = 2.84
Molecular weight: 96.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
850 |
850 |
850 |
850.0 |
96.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.353 ± 0.0 | 2.118 ± 0.0 |
6.471 ± 0.0 | 6.353 ± 0.0 |
4.235 ± 0.0 | 6.353 ± 0.0 |
2.353 ± 0.0 | 4.471 ± 0.0 |
6.471 ± 0.0 | 10.0 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.765 ± 0.0 | 2.235 ± 0.0 |
5.176 ± 0.0 | 3.176 ± 0.0 |
6.706 ± 0.0 | 8.235 ± 0.0 |
4.706 ± 0.0 | 6.588 ± 0.0 |
2.235 ± 0.0 | 4.0 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
