
Haloplanus vescus
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Haloferacaceae; Haloplanus
Average proteome isoelectric point is 4.9
Get precalculated fractions of proteins
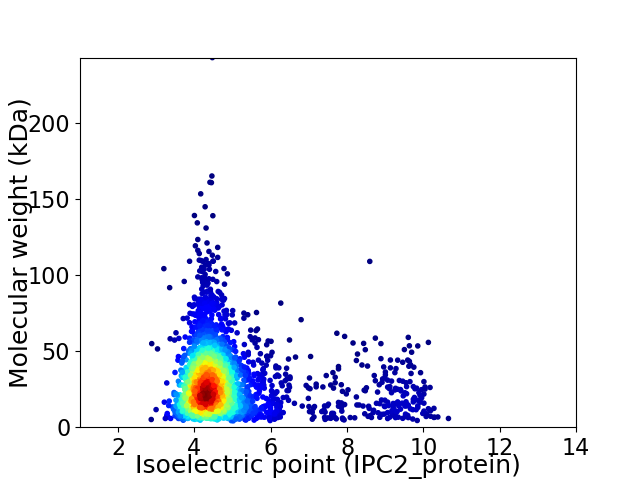
Virtual 2D-PAGE plot for 2791 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3VKD8|A0A1H3VKD8_9EURY ABC-2 type transport system ATP-binding protein OS=Haloplanus vescus OX=555874 GN=SAMN04488065_0039 PE=4 SV=1
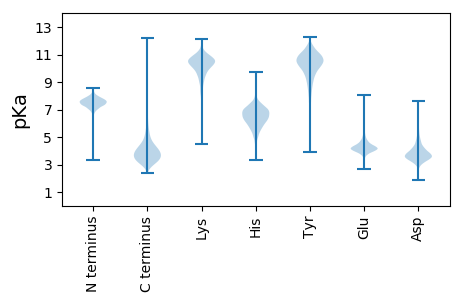
MM1 pKa = 7.61TDD3 pKa = 3.11YY4 pKa = 10.67NGKK7 pKa = 9.14VRR9 pKa = 11.84AVVLAALMVFSVFAGTVALSGTAAANVSSISGQSAQDD46 pKa = 3.39LVAGQTSVTQEE57 pKa = 3.71VTYY60 pKa = 9.95STTIPGDD67 pKa = 3.83TTPEE71 pKa = 4.36TYY73 pKa = 10.3TLDD76 pKa = 3.37ADD78 pKa = 4.08AYY80 pKa = 8.92EE81 pKa = 4.81GTSSATIQDD90 pKa = 3.4VSVSLRR96 pKa = 11.84NADD99 pKa = 4.14DD100 pKa = 3.88EE101 pKa = 5.17LSVGTVSEE109 pKa = 4.22SSGDD113 pKa = 3.76YY114 pKa = 11.05NIDD117 pKa = 3.41IQDD120 pKa = 3.73SGTDD124 pKa = 3.52GTTHH128 pKa = 7.13SFTLVLTITQDD139 pKa = 3.25TQGASTGTYY148 pKa = 10.92DD149 pKa = 3.82LTLSAASGASSTVSFDD165 pKa = 3.04ITSLSSNSDD174 pKa = 2.78KK175 pKa = 11.01RR176 pKa = 11.84AGNAAGSGTFDD187 pKa = 3.21TYY189 pKa = 11.33DD190 pKa = 3.48GEE192 pKa = 5.34GYY194 pKa = 10.94VFDD197 pKa = 4.77GANVYY202 pKa = 10.27QGEE205 pKa = 4.23SDD207 pKa = 3.57IEE209 pKa = 4.24LGGSISGGVVKK220 pKa = 9.29TAGNDD225 pKa = 3.16EE226 pKa = 4.34GVTLEE231 pKa = 4.92VPDD234 pKa = 4.63IPQDD238 pKa = 3.56QATGRR243 pKa = 11.84YY244 pKa = 5.35TTNGSNNAPGVTVQRR259 pKa = 11.84PRR261 pKa = 11.84VTTLDD266 pKa = 3.27VNNQYY271 pKa = 10.97GG272 pKa = 3.28
MM1 pKa = 7.61TDD3 pKa = 3.11YY4 pKa = 10.67NGKK7 pKa = 9.14VRR9 pKa = 11.84AVVLAALMVFSVFAGTVALSGTAAANVSSISGQSAQDD46 pKa = 3.39LVAGQTSVTQEE57 pKa = 3.71VTYY60 pKa = 9.95STTIPGDD67 pKa = 3.83TTPEE71 pKa = 4.36TYY73 pKa = 10.3TLDD76 pKa = 3.37ADD78 pKa = 4.08AYY80 pKa = 8.92EE81 pKa = 4.81GTSSATIQDD90 pKa = 3.4VSVSLRR96 pKa = 11.84NADD99 pKa = 4.14DD100 pKa = 3.88EE101 pKa = 5.17LSVGTVSEE109 pKa = 4.22SSGDD113 pKa = 3.76YY114 pKa = 11.05NIDD117 pKa = 3.41IQDD120 pKa = 3.73SGTDD124 pKa = 3.52GTTHH128 pKa = 7.13SFTLVLTITQDD139 pKa = 3.25TQGASTGTYY148 pKa = 10.92DD149 pKa = 3.82LTLSAASGASSTVSFDD165 pKa = 3.04ITSLSSNSDD174 pKa = 2.78KK175 pKa = 11.01RR176 pKa = 11.84AGNAAGSGTFDD187 pKa = 3.21TYY189 pKa = 11.33DD190 pKa = 3.48GEE192 pKa = 5.34GYY194 pKa = 10.94VFDD197 pKa = 4.77GANVYY202 pKa = 10.27QGEE205 pKa = 4.23SDD207 pKa = 3.57IEE209 pKa = 4.24LGGSISGGVVKK220 pKa = 9.29TAGNDD225 pKa = 3.16EE226 pKa = 4.34GVTLEE231 pKa = 4.92VPDD234 pKa = 4.63IPQDD238 pKa = 3.56QATGRR243 pKa = 11.84YY244 pKa = 5.35TTNGSNNAPGVTVQRR259 pKa = 11.84PRR261 pKa = 11.84VTTLDD266 pKa = 3.27VNNQYY271 pKa = 10.97GG272 pKa = 3.28
Molecular weight: 27.77 kDa
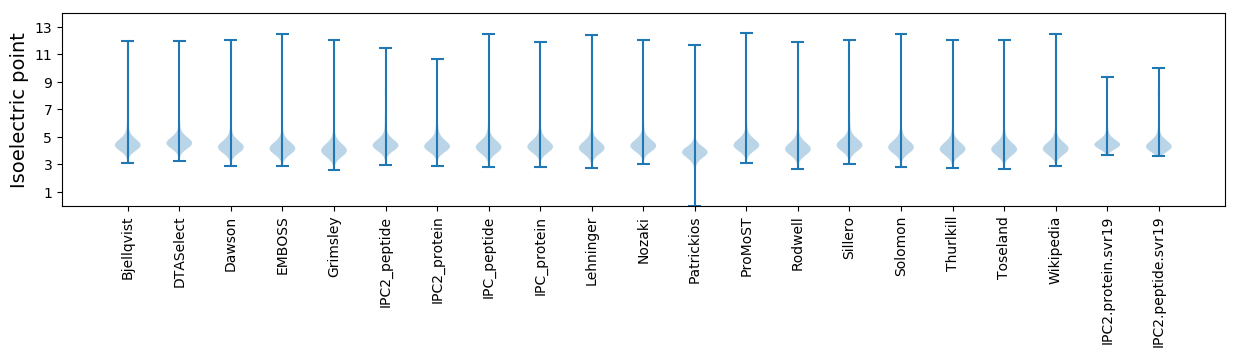
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H3WZD7|A0A1H3WZD7_9EURY Exonuclease III OS=Haloplanus vescus OX=555874 GN=SAMN04488065_1255 PE=4 SV=1
MM1 pKa = 7.52AAGSVALAAGLDD13 pKa = 3.68RR14 pKa = 11.84LVAEE18 pKa = 5.4PPAALHH24 pKa = 5.41PVAWLGRR31 pKa = 11.84ALGPFDD37 pKa = 5.0RR38 pKa = 11.84EE39 pKa = 3.81WRR41 pKa = 11.84HH42 pKa = 5.6PRR44 pKa = 11.84LAGAAVALAVPLFVAAAAAAVVTLGARR71 pKa = 11.84LDD73 pKa = 3.72PRR75 pKa = 11.84IGAVIAGLGCFAATSRR91 pKa = 11.84RR92 pKa = 11.84MLLDD96 pKa = 3.09EE97 pKa = 4.52ARR99 pKa = 11.84AVVAASDD106 pKa = 3.89TDD108 pKa = 4.07LSAARR113 pKa = 11.84TRR115 pKa = 11.84LRR117 pKa = 11.84ALAGRR122 pKa = 11.84DD123 pKa = 3.59ATDD126 pKa = 3.5LDD128 pKa = 3.99AGEE131 pKa = 4.28VRR133 pKa = 11.84SAAVEE138 pKa = 4.09SAAEE142 pKa = 3.98NLSDD146 pKa = 4.7GLVAPLAGFALFAPISLPAAAGAAAWVKK174 pKa = 10.46AVNTLDD180 pKa = 3.32STFGYY185 pKa = 10.3RR186 pKa = 11.84SIPMGWAPARR196 pKa = 11.84LDD198 pKa = 4.94DD199 pKa = 3.97IVMWVPARR207 pKa = 11.84VSAALLAVVAGRR219 pKa = 11.84PAALRR224 pKa = 11.84EE225 pKa = 4.02ARR227 pKa = 11.84SFARR231 pKa = 11.84RR232 pKa = 11.84PASPNAGWPMATLAVVLGVRR252 pKa = 11.84LTKK255 pKa = 10.57HH256 pKa = 5.26GAYY259 pKa = 10.18DD260 pKa = 3.79LRR262 pKa = 11.84PSASHH267 pKa = 5.91PTPAEE272 pKa = 3.81AEE274 pKa = 3.93RR275 pKa = 11.84GVAVVSRR282 pKa = 11.84AGWLAFGLTGVVALCC297 pKa = 4.42
MM1 pKa = 7.52AAGSVALAAGLDD13 pKa = 3.68RR14 pKa = 11.84LVAEE18 pKa = 5.4PPAALHH24 pKa = 5.41PVAWLGRR31 pKa = 11.84ALGPFDD37 pKa = 5.0RR38 pKa = 11.84EE39 pKa = 3.81WRR41 pKa = 11.84HH42 pKa = 5.6PRR44 pKa = 11.84LAGAAVALAVPLFVAAAAAAVVTLGARR71 pKa = 11.84LDD73 pKa = 3.72PRR75 pKa = 11.84IGAVIAGLGCFAATSRR91 pKa = 11.84RR92 pKa = 11.84MLLDD96 pKa = 3.09EE97 pKa = 4.52ARR99 pKa = 11.84AVVAASDD106 pKa = 3.89TDD108 pKa = 4.07LSAARR113 pKa = 11.84TRR115 pKa = 11.84LRR117 pKa = 11.84ALAGRR122 pKa = 11.84DD123 pKa = 3.59ATDD126 pKa = 3.5LDD128 pKa = 3.99AGEE131 pKa = 4.28VRR133 pKa = 11.84SAAVEE138 pKa = 4.09SAAEE142 pKa = 3.98NLSDD146 pKa = 4.7GLVAPLAGFALFAPISLPAAAGAAAWVKK174 pKa = 10.46AVNTLDD180 pKa = 3.32STFGYY185 pKa = 10.3RR186 pKa = 11.84SIPMGWAPARR196 pKa = 11.84LDD198 pKa = 4.94DD199 pKa = 3.97IVMWVPARR207 pKa = 11.84VSAALLAVVAGRR219 pKa = 11.84PAALRR224 pKa = 11.84EE225 pKa = 4.02ARR227 pKa = 11.84SFARR231 pKa = 11.84RR232 pKa = 11.84PASPNAGWPMATLAVVLGVRR252 pKa = 11.84LTKK255 pKa = 10.57HH256 pKa = 5.26GAYY259 pKa = 10.18DD260 pKa = 3.79LRR262 pKa = 11.84PSASHH267 pKa = 5.91PTPAEE272 pKa = 3.81AEE274 pKa = 3.93RR275 pKa = 11.84GVAVVSRR282 pKa = 11.84AGWLAFGLTGVVALCC297 pKa = 4.42
Molecular weight: 30.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
803080 |
39 |
2172 |
287.7 |
31.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.121 ± 0.068 | 0.705 ± 0.015 |
8.884 ± 0.065 | 8.047 ± 0.069 |
3.245 ± 0.032 | 8.263 ± 0.052 |
2.021 ± 0.022 | 3.864 ± 0.034 |
1.706 ± 0.026 | 9.066 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.745 ± 0.022 | 2.235 ± 0.023 |
4.669 ± 0.032 | 2.491 ± 0.029 |
6.789 ± 0.048 | 5.463 ± 0.041 |
6.61 ± 0.041 | 9.305 ± 0.054 |
1.114 ± 0.017 | 2.656 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
