
Bos taurus papillomavirus 12
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Xipapillomavirus; Xipapillomavirus 2
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
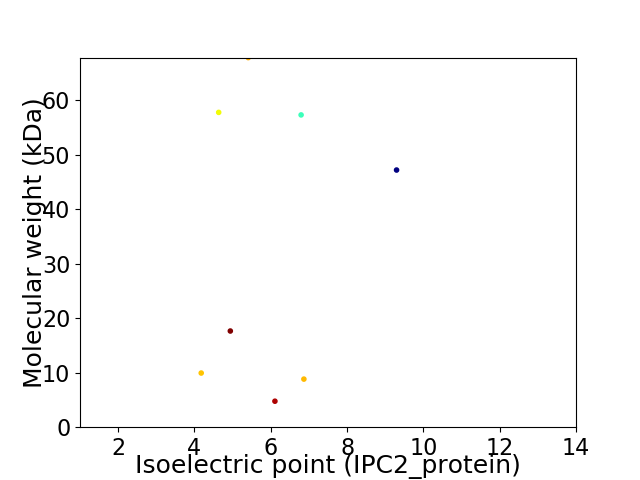
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G1CR70|G1CR70_9PAPI Replication protein E1 OS=Bos taurus papillomavirus 12 OX=1070324 GN=E1 PE=3 SV=1
MM1 pKa = 7.82RR2 pKa = 11.84GSQPAISDD10 pKa = 3.1ISEE13 pKa = 4.0EE14 pKa = 4.12LQEE17 pKa = 4.37LVCPVNLEE25 pKa = 4.07CDD27 pKa = 3.41EE28 pKa = 4.31EE29 pKa = 4.15LAPEE33 pKa = 4.3NDD35 pKa = 3.58DD36 pKa = 4.04PYY38 pKa = 11.56AVAVACHH45 pKa = 6.0KK46 pKa = 10.58CGNQVAFCVYY56 pKa = 10.47APRR59 pKa = 11.84EE60 pKa = 4.38GIRR63 pKa = 11.84QFQQLLLDD71 pKa = 5.2CLDD74 pKa = 4.43FCCPACSLNLLQRR87 pKa = 11.84NGLL90 pKa = 3.69
MM1 pKa = 7.82RR2 pKa = 11.84GSQPAISDD10 pKa = 3.1ISEE13 pKa = 4.0EE14 pKa = 4.12LQEE17 pKa = 4.37LVCPVNLEE25 pKa = 4.07CDD27 pKa = 3.41EE28 pKa = 4.31EE29 pKa = 4.15LAPEE33 pKa = 4.3NDD35 pKa = 3.58DD36 pKa = 4.04PYY38 pKa = 11.56AVAVACHH45 pKa = 6.0KK46 pKa = 10.58CGNQVAFCVYY56 pKa = 10.47APRR59 pKa = 11.84EE60 pKa = 4.38GIRR63 pKa = 11.84QFQQLLLDD71 pKa = 5.2CLDD74 pKa = 4.43FCCPACSLNLLQRR87 pKa = 11.84NGLL90 pKa = 3.69
Molecular weight: 9.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G1CR72|G1CR72_9PAPI E4 protein OS=Bos taurus papillomavirus 12 OX=1070324 GN=E4 PE=4 SV=1
MM1 pKa = 8.04DD2 pKa = 5.48PLAARR7 pKa = 11.84FDD9 pKa = 3.95ALQEE13 pKa = 3.89EE14 pKa = 5.03LMQIYY19 pKa = 10.06EE20 pKa = 4.29RR21 pKa = 11.84GEE23 pKa = 4.18STLDD27 pKa = 3.01AQIRR31 pKa = 11.84HH32 pKa = 5.8WEE34 pKa = 4.42LIRR37 pKa = 11.84HH38 pKa = 5.18EE39 pKa = 4.08QATYY43 pKa = 9.83FVARR47 pKa = 11.84RR48 pKa = 11.84QGITRR53 pKa = 11.84LGLYY57 pKa = 7.23PVPSASVSEE66 pKa = 4.28ARR68 pKa = 11.84AKK70 pKa = 10.02QAIEE74 pKa = 3.75MHH76 pKa = 6.88LLLQSLKK83 pKa = 10.15KK84 pKa = 10.13SAYY87 pKa = 9.86ADD89 pKa = 3.68EE90 pKa = 4.63RR91 pKa = 11.84WTLVEE96 pKa = 3.87TSFEE100 pKa = 4.41HH101 pKa = 6.78FNSPPARR108 pKa = 11.84TLKK111 pKa = 10.55KK112 pKa = 10.49GPTQVTVIYY121 pKa = 10.62DD122 pKa = 3.89NNPEE126 pKa = 3.86NAMTYY131 pKa = 7.21TLWKK135 pKa = 9.07YY136 pKa = 11.23VYY138 pKa = 10.63VLDD141 pKa = 5.73PNDD144 pKa = 3.32VWHH147 pKa = 7.05KK148 pKa = 10.66LPSDD152 pKa = 3.0VDD154 pKa = 3.82YY155 pKa = 11.77YY156 pKa = 11.33GIYY159 pKa = 8.9YY160 pKa = 9.65TDD162 pKa = 4.66LDD164 pKa = 3.86QQRR167 pKa = 11.84VYY169 pKa = 10.64YY170 pKa = 10.77VSFGTDD176 pKa = 2.73AEE178 pKa = 4.53QYY180 pKa = 8.91TGSGQWTVHH189 pKa = 5.92FANKK193 pKa = 8.69TFSAPVTSSAGGHH206 pKa = 5.9SGLEE210 pKa = 4.15EE211 pKa = 3.95EE212 pKa = 4.73QAQGPRR218 pKa = 11.84QLAARR223 pKa = 11.84RR224 pKa = 11.84QATYY228 pKa = 9.74RR229 pKa = 11.84GRR231 pKa = 11.84RR232 pKa = 11.84SRR234 pKa = 11.84RR235 pKa = 11.84SRR237 pKa = 11.84TPQTPPRR244 pKa = 11.84SRR246 pKa = 11.84SRR248 pKa = 11.84SEE250 pKa = 3.63SRR252 pKa = 11.84SRR254 pKa = 11.84SASSSSDD261 pKa = 2.85TDD263 pKa = 2.95HH264 pKa = 7.78RR265 pKa = 11.84SRR267 pKa = 11.84SRR269 pKa = 11.84QRR271 pKa = 11.84NGGRR275 pKa = 11.84GFRR278 pKa = 11.84RR279 pKa = 11.84GDD281 pKa = 3.24STGEE285 pKa = 3.87HH286 pKa = 6.89PEE288 pKa = 3.96SEE290 pKa = 4.6EE291 pKa = 3.53EE292 pKa = 4.22SGRR295 pKa = 11.84HH296 pKa = 4.87SPGPPGSLGRR306 pKa = 11.84RR307 pKa = 11.84TAEE310 pKa = 3.69TSGASGRR317 pKa = 11.84RR318 pKa = 11.84RR319 pKa = 11.84TPRR322 pKa = 11.84VVQLLKK328 pKa = 10.83DD329 pKa = 3.52AADD332 pKa = 3.95PPVLLLRR339 pKa = 11.84GPANTLKK346 pKa = 10.7GFRR349 pKa = 11.84RR350 pKa = 11.84KK351 pKa = 9.81ARR353 pKa = 11.84LKK355 pKa = 8.39YY356 pKa = 10.0SEE358 pKa = 5.27YY359 pKa = 9.32YY360 pKa = 10.72SCMSTAFSWVSGTCTEE376 pKa = 5.04RR377 pKa = 11.84LGTCQRR383 pKa = 11.84MLVAFNSDD391 pKa = 3.23SQRR394 pKa = 11.84TRR396 pKa = 11.84FLCNVHH402 pKa = 6.47IPRR405 pKa = 11.84SVTFVKK411 pKa = 10.85GSFDD415 pKa = 3.5ALL417 pKa = 3.54
MM1 pKa = 8.04DD2 pKa = 5.48PLAARR7 pKa = 11.84FDD9 pKa = 3.95ALQEE13 pKa = 3.89EE14 pKa = 5.03LMQIYY19 pKa = 10.06EE20 pKa = 4.29RR21 pKa = 11.84GEE23 pKa = 4.18STLDD27 pKa = 3.01AQIRR31 pKa = 11.84HH32 pKa = 5.8WEE34 pKa = 4.42LIRR37 pKa = 11.84HH38 pKa = 5.18EE39 pKa = 4.08QATYY43 pKa = 9.83FVARR47 pKa = 11.84RR48 pKa = 11.84QGITRR53 pKa = 11.84LGLYY57 pKa = 7.23PVPSASVSEE66 pKa = 4.28ARR68 pKa = 11.84AKK70 pKa = 10.02QAIEE74 pKa = 3.75MHH76 pKa = 6.88LLLQSLKK83 pKa = 10.15KK84 pKa = 10.13SAYY87 pKa = 9.86ADD89 pKa = 3.68EE90 pKa = 4.63RR91 pKa = 11.84WTLVEE96 pKa = 3.87TSFEE100 pKa = 4.41HH101 pKa = 6.78FNSPPARR108 pKa = 11.84TLKK111 pKa = 10.55KK112 pKa = 10.49GPTQVTVIYY121 pKa = 10.62DD122 pKa = 3.89NNPEE126 pKa = 3.86NAMTYY131 pKa = 7.21TLWKK135 pKa = 9.07YY136 pKa = 11.23VYY138 pKa = 10.63VLDD141 pKa = 5.73PNDD144 pKa = 3.32VWHH147 pKa = 7.05KK148 pKa = 10.66LPSDD152 pKa = 3.0VDD154 pKa = 3.82YY155 pKa = 11.77YY156 pKa = 11.33GIYY159 pKa = 8.9YY160 pKa = 9.65TDD162 pKa = 4.66LDD164 pKa = 3.86QQRR167 pKa = 11.84VYY169 pKa = 10.64YY170 pKa = 10.77VSFGTDD176 pKa = 2.73AEE178 pKa = 4.53QYY180 pKa = 8.91TGSGQWTVHH189 pKa = 5.92FANKK193 pKa = 8.69TFSAPVTSSAGGHH206 pKa = 5.9SGLEE210 pKa = 4.15EE211 pKa = 3.95EE212 pKa = 4.73QAQGPRR218 pKa = 11.84QLAARR223 pKa = 11.84RR224 pKa = 11.84QATYY228 pKa = 9.74RR229 pKa = 11.84GRR231 pKa = 11.84RR232 pKa = 11.84SRR234 pKa = 11.84RR235 pKa = 11.84SRR237 pKa = 11.84TPQTPPRR244 pKa = 11.84SRR246 pKa = 11.84SRR248 pKa = 11.84SEE250 pKa = 3.63SRR252 pKa = 11.84SRR254 pKa = 11.84SASSSSDD261 pKa = 2.85TDD263 pKa = 2.95HH264 pKa = 7.78RR265 pKa = 11.84SRR267 pKa = 11.84SRR269 pKa = 11.84QRR271 pKa = 11.84NGGRR275 pKa = 11.84GFRR278 pKa = 11.84RR279 pKa = 11.84GDD281 pKa = 3.24STGEE285 pKa = 3.87HH286 pKa = 6.89PEE288 pKa = 3.96SEE290 pKa = 4.6EE291 pKa = 3.53EE292 pKa = 4.22SGRR295 pKa = 11.84HH296 pKa = 4.87SPGPPGSLGRR306 pKa = 11.84RR307 pKa = 11.84TAEE310 pKa = 3.69TSGASGRR317 pKa = 11.84RR318 pKa = 11.84RR319 pKa = 11.84TPRR322 pKa = 11.84VVQLLKK328 pKa = 10.83DD329 pKa = 3.52AADD332 pKa = 3.95PPVLLLRR339 pKa = 11.84GPANTLKK346 pKa = 10.7GFRR349 pKa = 11.84RR350 pKa = 11.84KK351 pKa = 9.81ARR353 pKa = 11.84LKK355 pKa = 8.39YY356 pKa = 10.0SEE358 pKa = 5.27YY359 pKa = 9.32YY360 pKa = 10.72SCMSTAFSWVSGTCTEE376 pKa = 5.04RR377 pKa = 11.84LGTCQRR383 pKa = 11.84MLVAFNSDD391 pKa = 3.23SQRR394 pKa = 11.84TRR396 pKa = 11.84FLCNVHH402 pKa = 6.47IPRR405 pKa = 11.84SVTFVKK411 pKa = 10.85GSFDD415 pKa = 3.5ALL417 pKa = 3.54
Molecular weight: 47.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2409 |
41 |
596 |
301.1 |
33.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.853 ± 0.684 | 1.868 ± 0.546 |
6.476 ± 0.649 | 6.185 ± 0.357 |
4.608 ± 0.491 | 6.434 ± 0.689 |
1.91 ± 0.184 | 4.4 ± 0.735 |
4.649 ± 0.969 | 9.174 ± 1.352 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.743 ± 0.225 | 4.525 ± 0.666 |
6.517 ± 0.8 | 4.774 ± 0.291 |
6.434 ± 1.078 | 7.057 ± 0.815 |
6.517 ± 0.787 | 6.31 ± 0.484 |
1.411 ± 0.341 | 3.155 ± 0.34 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
