
Pseudoroseicyclus sp. CLL3-39
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Pseudoroseicyclus
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
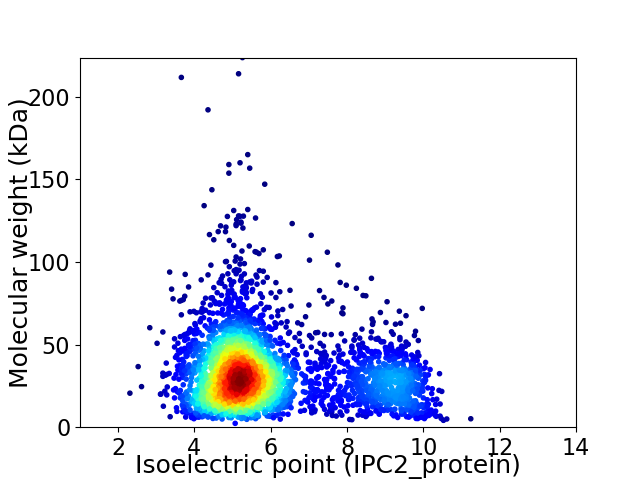
Virtual 2D-PAGE plot for 3556 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
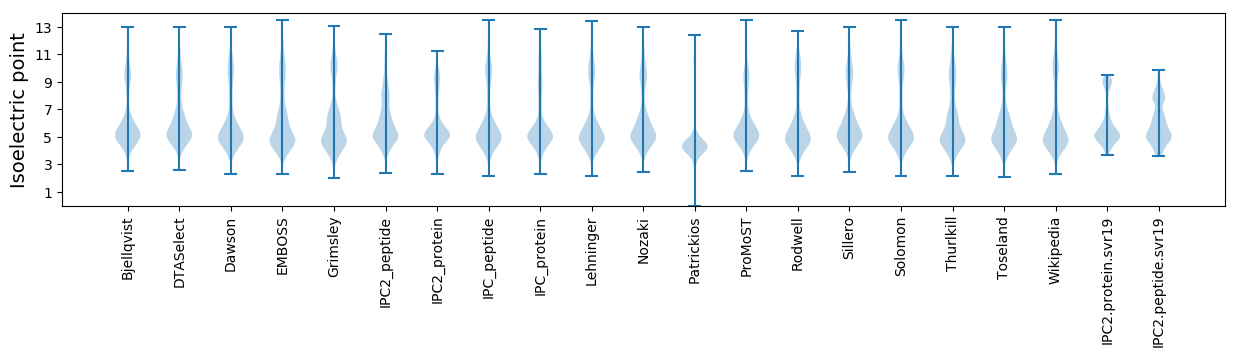
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6B2JUS4|A0A6B2JUS4_9RHOB Uncharacterized protein OS=Pseudoroseicyclus sp. CLL3-39 OX=2705421 GN=GZA08_02975 PE=4 SV=1
MM1 pKa = 8.12DD2 pKa = 6.09DD3 pKa = 3.82EE4 pKa = 5.07AADD7 pKa = 3.9TATDD11 pKa = 3.55EE12 pKa = 5.2APTLDD17 pKa = 3.56EE18 pKa = 4.51VEE20 pKa = 4.63PGEE23 pKa = 4.8DD24 pKa = 3.18ALFVDD29 pKa = 5.21DD30 pKa = 6.48LEE32 pKa = 4.63TATDD36 pKa = 3.96DD37 pKa = 4.0EE38 pKa = 5.19PGAEE42 pKa = 3.8ATASEE47 pKa = 4.81EE48 pKa = 4.07EE49 pKa = 4.4SLDD52 pKa = 3.87AVIDD56 pKa = 3.92PAPEE60 pKa = 3.85TGEE63 pKa = 3.96TSEE66 pKa = 4.74APEE69 pKa = 5.02DD70 pKa = 4.09GPAATEE76 pKa = 3.94PEE78 pKa = 4.21PVFEE82 pKa = 5.83VGADD86 pKa = 3.11WPYY89 pKa = 10.58WGGDD93 pKa = 3.16ADD95 pKa = 3.84ATRR98 pKa = 11.84YY99 pKa = 10.33SPLAQITPQNVEE111 pKa = 3.83EE112 pKa = 4.0LVEE115 pKa = 4.2VFSYY119 pKa = 8.99QTGDD123 pKa = 3.05MPTEE127 pKa = 4.26VTEE130 pKa = 4.34GTYY133 pKa = 10.58SPEE136 pKa = 3.91TTPLKK141 pKa = 10.95LGDD144 pKa = 4.42DD145 pKa = 4.46LLMCSAMNILISIDD159 pKa = 3.28AATGEE164 pKa = 4.52EE165 pKa = 3.76NWRR168 pKa = 11.84FNPGVPPEE176 pKa = 4.77AIPYY180 pKa = 8.85GATCRR185 pKa = 11.84GVSVYY190 pKa = 10.4RR191 pKa = 11.84DD192 pKa = 3.42PDD194 pKa = 3.45AGEE197 pKa = 4.84GDD199 pKa = 3.72LCATRR204 pKa = 11.84VIQATLDD211 pKa = 3.56AKK213 pKa = 10.71LVAVDD218 pKa = 4.75AEE220 pKa = 4.51TGLPCEE226 pKa = 4.74DD227 pKa = 3.91FGEE230 pKa = 4.41NGTVDD235 pKa = 3.53LWQDD239 pKa = 2.44IGEE242 pKa = 4.63RR243 pKa = 11.84VPGWYY248 pKa = 10.17AVTAPPAIVRR258 pKa = 11.84GVVVTGAQVLDD269 pKa = 4.32GQAEE273 pKa = 4.08EE274 pKa = 4.56APSGVVRR281 pKa = 11.84GYY283 pKa = 10.95DD284 pKa = 3.16ALTGEE289 pKa = 5.21LAWAWDD295 pKa = 3.58LAAPEE300 pKa = 4.33EE301 pKa = 4.49MRR303 pKa = 11.84DD304 pKa = 3.74GPPEE308 pKa = 3.82GRR310 pKa = 11.84VYY312 pKa = 10.87TRR314 pKa = 11.84GTPNMWTTAVADD326 pKa = 3.84EE327 pKa = 4.26EE328 pKa = 4.35LGYY331 pKa = 11.16VYY333 pKa = 10.77LPLGNSSVDD342 pKa = 3.48YY343 pKa = 10.41YY344 pKa = 11.56GSNRR348 pKa = 11.84SEE350 pKa = 4.17AEE352 pKa = 4.06NEE354 pKa = 3.92FSTSLVAVDD363 pKa = 3.53ATTGEE368 pKa = 4.68EE369 pKa = 3.94VWHH372 pKa = 5.93FQTVRR377 pKa = 11.84RR378 pKa = 11.84DD379 pKa = 2.9VWDD382 pKa = 3.73YY383 pKa = 11.96DD384 pKa = 4.13LGSQPTLVDD393 pKa = 3.36WQGTPAIILPSKK405 pKa = 10.61QGDD408 pKa = 3.49IYY410 pKa = 10.89ILDD413 pKa = 4.02RR414 pKa = 11.84ATGEE418 pKa = 3.95PLTPVGQLEE427 pKa = 4.65DD428 pKa = 3.84LPQGEE433 pKa = 4.95IEE435 pKa = 4.45PDD437 pKa = 3.99YY438 pKa = 11.29ISDD441 pKa = 3.74TQPYY445 pKa = 9.0SVWHH449 pKa = 5.75TLRR452 pKa = 11.84KK453 pKa = 9.3PPLEE457 pKa = 5.22AKK459 pKa = 9.83DD460 pKa = 3.38AWGFTPIDD468 pKa = 3.74QLWCRR473 pKa = 11.84IQFHH477 pKa = 6.18RR478 pKa = 11.84ANYY481 pKa = 9.07EE482 pKa = 4.15GYY484 pKa = 7.72FTPPSADD491 pKa = 3.67APWIQYY497 pKa = 9.29PGYY500 pKa = 10.62NGGSDD505 pKa = 3.32WGSVAVDD512 pKa = 3.25VEE514 pKa = 4.25RR515 pKa = 11.84GIIIANYY522 pKa = 10.31NDD524 pKa = 3.23IPNYY528 pKa = 8.76NQLIEE533 pKa = 5.54RR534 pKa = 11.84EE535 pKa = 4.4DD536 pKa = 3.98PYY538 pKa = 10.92QTDD541 pKa = 3.79EE542 pKa = 4.17YY543 pKa = 9.11TTIDD547 pKa = 3.85EE548 pKa = 4.62APEE551 pKa = 4.45SVGSPADD558 pKa = 3.67NEE560 pKa = 4.46AGPQAGSPYY569 pKa = 10.06AISVNAGWRR578 pKa = 11.84SEE580 pKa = 4.06LTGMPCSSPPYY591 pKa = 9.69GGIRR595 pKa = 11.84AIDD598 pKa = 3.95LEE600 pKa = 4.43TGEE603 pKa = 4.58TLWDD607 pKa = 3.9RR608 pKa = 11.84PLGTARR614 pKa = 11.84RR615 pKa = 11.84NGPFGIPSFLPLDD628 pKa = 3.37IGTPNNGGSVVTASGLVFIGAATDD652 pKa = 3.48NLFRR656 pKa = 11.84AIDD659 pKa = 3.58IEE661 pKa = 4.32TGEE664 pKa = 4.45TLWSVVLPAGGQANPVTYY682 pKa = 10.12EE683 pKa = 3.57VDD685 pKa = 2.8GRR687 pKa = 11.84QYY689 pKa = 12.02VMISAMGHH697 pKa = 5.83HH698 pKa = 6.34FMEE701 pKa = 5.06TGVGDD706 pKa = 4.76YY707 pKa = 10.49MMAWALPEE715 pKa = 3.81
MM1 pKa = 8.12DD2 pKa = 6.09DD3 pKa = 3.82EE4 pKa = 5.07AADD7 pKa = 3.9TATDD11 pKa = 3.55EE12 pKa = 5.2APTLDD17 pKa = 3.56EE18 pKa = 4.51VEE20 pKa = 4.63PGEE23 pKa = 4.8DD24 pKa = 3.18ALFVDD29 pKa = 5.21DD30 pKa = 6.48LEE32 pKa = 4.63TATDD36 pKa = 3.96DD37 pKa = 4.0EE38 pKa = 5.19PGAEE42 pKa = 3.8ATASEE47 pKa = 4.81EE48 pKa = 4.07EE49 pKa = 4.4SLDD52 pKa = 3.87AVIDD56 pKa = 3.92PAPEE60 pKa = 3.85TGEE63 pKa = 3.96TSEE66 pKa = 4.74APEE69 pKa = 5.02DD70 pKa = 4.09GPAATEE76 pKa = 3.94PEE78 pKa = 4.21PVFEE82 pKa = 5.83VGADD86 pKa = 3.11WPYY89 pKa = 10.58WGGDD93 pKa = 3.16ADD95 pKa = 3.84ATRR98 pKa = 11.84YY99 pKa = 10.33SPLAQITPQNVEE111 pKa = 3.83EE112 pKa = 4.0LVEE115 pKa = 4.2VFSYY119 pKa = 8.99QTGDD123 pKa = 3.05MPTEE127 pKa = 4.26VTEE130 pKa = 4.34GTYY133 pKa = 10.58SPEE136 pKa = 3.91TTPLKK141 pKa = 10.95LGDD144 pKa = 4.42DD145 pKa = 4.46LLMCSAMNILISIDD159 pKa = 3.28AATGEE164 pKa = 4.52EE165 pKa = 3.76NWRR168 pKa = 11.84FNPGVPPEE176 pKa = 4.77AIPYY180 pKa = 8.85GATCRR185 pKa = 11.84GVSVYY190 pKa = 10.4RR191 pKa = 11.84DD192 pKa = 3.42PDD194 pKa = 3.45AGEE197 pKa = 4.84GDD199 pKa = 3.72LCATRR204 pKa = 11.84VIQATLDD211 pKa = 3.56AKK213 pKa = 10.71LVAVDD218 pKa = 4.75AEE220 pKa = 4.51TGLPCEE226 pKa = 4.74DD227 pKa = 3.91FGEE230 pKa = 4.41NGTVDD235 pKa = 3.53LWQDD239 pKa = 2.44IGEE242 pKa = 4.63RR243 pKa = 11.84VPGWYY248 pKa = 10.17AVTAPPAIVRR258 pKa = 11.84GVVVTGAQVLDD269 pKa = 4.32GQAEE273 pKa = 4.08EE274 pKa = 4.56APSGVVRR281 pKa = 11.84GYY283 pKa = 10.95DD284 pKa = 3.16ALTGEE289 pKa = 5.21LAWAWDD295 pKa = 3.58LAAPEE300 pKa = 4.33EE301 pKa = 4.49MRR303 pKa = 11.84DD304 pKa = 3.74GPPEE308 pKa = 3.82GRR310 pKa = 11.84VYY312 pKa = 10.87TRR314 pKa = 11.84GTPNMWTTAVADD326 pKa = 3.84EE327 pKa = 4.26EE328 pKa = 4.35LGYY331 pKa = 11.16VYY333 pKa = 10.77LPLGNSSVDD342 pKa = 3.48YY343 pKa = 10.41YY344 pKa = 11.56GSNRR348 pKa = 11.84SEE350 pKa = 4.17AEE352 pKa = 4.06NEE354 pKa = 3.92FSTSLVAVDD363 pKa = 3.53ATTGEE368 pKa = 4.68EE369 pKa = 3.94VWHH372 pKa = 5.93FQTVRR377 pKa = 11.84RR378 pKa = 11.84DD379 pKa = 2.9VWDD382 pKa = 3.73YY383 pKa = 11.96DD384 pKa = 4.13LGSQPTLVDD393 pKa = 3.36WQGTPAIILPSKK405 pKa = 10.61QGDD408 pKa = 3.49IYY410 pKa = 10.89ILDD413 pKa = 4.02RR414 pKa = 11.84ATGEE418 pKa = 3.95PLTPVGQLEE427 pKa = 4.65DD428 pKa = 3.84LPQGEE433 pKa = 4.95IEE435 pKa = 4.45PDD437 pKa = 3.99YY438 pKa = 11.29ISDD441 pKa = 3.74TQPYY445 pKa = 9.0SVWHH449 pKa = 5.75TLRR452 pKa = 11.84KK453 pKa = 9.3PPLEE457 pKa = 5.22AKK459 pKa = 9.83DD460 pKa = 3.38AWGFTPIDD468 pKa = 3.74QLWCRR473 pKa = 11.84IQFHH477 pKa = 6.18RR478 pKa = 11.84ANYY481 pKa = 9.07EE482 pKa = 4.15GYY484 pKa = 7.72FTPPSADD491 pKa = 3.67APWIQYY497 pKa = 9.29PGYY500 pKa = 10.62NGGSDD505 pKa = 3.32WGSVAVDD512 pKa = 3.25VEE514 pKa = 4.25RR515 pKa = 11.84GIIIANYY522 pKa = 10.31NDD524 pKa = 3.23IPNYY528 pKa = 8.76NQLIEE533 pKa = 5.54RR534 pKa = 11.84EE535 pKa = 4.4DD536 pKa = 3.98PYY538 pKa = 10.92QTDD541 pKa = 3.79EE542 pKa = 4.17YY543 pKa = 9.11TTIDD547 pKa = 3.85EE548 pKa = 4.62APEE551 pKa = 4.45SVGSPADD558 pKa = 3.67NEE560 pKa = 4.46AGPQAGSPYY569 pKa = 10.06AISVNAGWRR578 pKa = 11.84SEE580 pKa = 4.06LTGMPCSSPPYY591 pKa = 9.69GGIRR595 pKa = 11.84AIDD598 pKa = 3.95LEE600 pKa = 4.43TGEE603 pKa = 4.58TLWDD607 pKa = 3.9RR608 pKa = 11.84PLGTARR614 pKa = 11.84RR615 pKa = 11.84NGPFGIPSFLPLDD628 pKa = 3.37IGTPNNGGSVVTASGLVFIGAATDD652 pKa = 3.48NLFRR656 pKa = 11.84AIDD659 pKa = 3.58IEE661 pKa = 4.32TGEE664 pKa = 4.45TLWSVVLPAGGQANPVTYY682 pKa = 10.12EE683 pKa = 3.57VDD685 pKa = 2.8GRR687 pKa = 11.84QYY689 pKa = 12.02VMISAMGHH697 pKa = 5.83HH698 pKa = 6.34FMEE701 pKa = 5.06TGVGDD706 pKa = 4.76YY707 pKa = 10.49MMAWALPEE715 pKa = 3.81
Molecular weight: 77.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6B2JLW5|A0A6B2JLW5_9RHOB Response regulator transcription factor OS=Pseudoroseicyclus sp. CLL3-39 OX=2705421 GN=GZA08_16455 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 9.2ARR15 pKa = 11.84HH16 pKa = 4.71GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.97VLSAA44 pKa = 4.11
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 9.2ARR15 pKa = 11.84HH16 pKa = 4.71GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.97VLSAA44 pKa = 4.11
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1122436 |
21 |
2097 |
315.6 |
34.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.483 ± 0.065 | 0.786 ± 0.013 |
5.482 ± 0.038 | 6.893 ± 0.039 |
3.517 ± 0.024 | 9.302 ± 0.046 |
1.994 ± 0.019 | 4.709 ± 0.031 |
2.319 ± 0.034 | 10.475 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.628 ± 0.02 | 2.094 ± 0.02 |
5.659 ± 0.036 | 2.818 ± 0.023 |
7.157 ± 0.047 | 4.925 ± 0.026 |
5.174 ± 0.026 | 6.992 ± 0.036 |
1.471 ± 0.02 | 2.123 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
