
Flavobacterium phage Fpv6
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
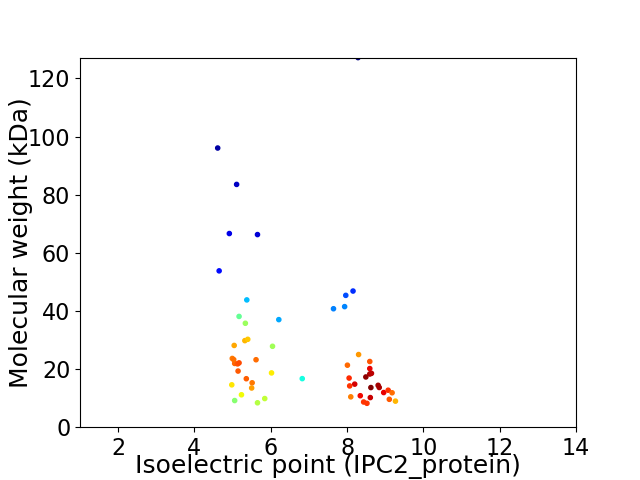
Virtual 2D-PAGE plot for 58 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B0WLD3|A0A1B0WLD3_9CAUD Uncharacterized protein OS=Flavobacterium phage Fpv6 OX=1814286 PE=4 SV=1
MM1 pKa = 7.12VKK3 pKa = 10.44VIEE6 pKa = 4.15RR7 pKa = 11.84TYY9 pKa = 11.5NEE11 pKa = 3.89IFTPEE16 pKa = 3.98STSWLLGNVGEE27 pKa = 4.19WQKK30 pKa = 11.54LKK32 pKa = 11.1LKK34 pKa = 10.9VEE36 pKa = 4.22VSVDD40 pKa = 3.87LYY42 pKa = 11.17CDD44 pKa = 3.07QEE46 pKa = 4.01NRR48 pKa = 11.84LKK50 pKa = 10.18IDD52 pKa = 3.65YY53 pKa = 10.12INNSFVLNDD62 pKa = 3.51GKK64 pKa = 10.87KK65 pKa = 8.31WSDD68 pKa = 3.23YY69 pKa = 11.43GFDD72 pKa = 3.18IGQNIQLVYY81 pKa = 10.64KK82 pKa = 10.43FKK84 pKa = 11.14LDD86 pKa = 3.63TNGDD90 pKa = 4.01GIYY93 pKa = 10.22EE94 pKa = 4.37GPFYY98 pKa = 10.2QAPNFTIVNIASNVMEE114 pKa = 4.24VLQPIVIWVLADD126 pKa = 4.1TIPYY130 pKa = 8.93NQDD133 pKa = 2.34SVRR136 pKa = 11.84IIDD139 pKa = 3.77CKK141 pKa = 10.69FIAFVEE147 pKa = 4.36PEE149 pKa = 4.07GCRR152 pKa = 11.84IVYY155 pKa = 8.92GQISNEE161 pKa = 3.71NSEE164 pKa = 4.3NLSLQSFIDD173 pKa = 3.62GTTTMFVQPNVKK185 pKa = 9.93SQSLNINSSMNAVGMQSGMSVRR207 pKa = 11.84MVSVKK212 pKa = 10.44RR213 pKa = 11.84IGLKK217 pKa = 10.16PGSQNVYY224 pKa = 9.29QYY226 pKa = 11.08EE227 pKa = 4.12FEE229 pKa = 4.18LQYY232 pKa = 10.96LISSFFEE239 pKa = 4.43NISNFTNNQMPDD251 pKa = 3.37YY252 pKa = 10.39LLGDD256 pKa = 4.07GSLTDD261 pKa = 3.45NFQILFYY268 pKa = 10.36PKK270 pKa = 9.29WNNPNTFIQNDD281 pKa = 3.62MKK283 pKa = 10.43KK284 pKa = 8.0TARR287 pKa = 11.84LGNTGWFDD295 pKa = 3.56EE296 pKa = 4.86NYY298 pKa = 10.94NEE300 pKa = 4.82LEE302 pKa = 4.08NNFVIEE308 pKa = 4.06SLQYY312 pKa = 10.81FDD314 pKa = 6.38DD315 pKa = 4.01NGNPVDD321 pKa = 5.06AIDD324 pKa = 3.74YY325 pKa = 7.49FAPTKK330 pKa = 10.82VKK332 pKa = 10.74GIVSGVQNINTNTEE346 pKa = 3.97CGFGFIWVPKK356 pKa = 10.4FEE358 pKa = 4.5EE359 pKa = 4.73DD360 pKa = 3.91YY361 pKa = 11.21KK362 pKa = 11.42NKK364 pKa = 7.82EE365 pKa = 3.98TPFYY369 pKa = 11.18QNTFVNTGTTTDD381 pKa = 3.39GFKK384 pKa = 10.91VGTFYY389 pKa = 11.07PLTNVGGGLNSASIDD404 pKa = 3.57TKK406 pKa = 10.55NVKK409 pKa = 8.64FTDD412 pKa = 3.03IGGGKK417 pKa = 9.35ISFEE421 pKa = 4.52MIFTPNPNFYY431 pKa = 10.93LFFDD435 pKa = 4.38ARR437 pKa = 11.84NDD439 pKa = 3.71DD440 pKa = 3.53DD441 pKa = 4.51RR442 pKa = 11.84KK443 pKa = 9.43YY444 pKa = 11.16VLWLSVADD452 pKa = 3.6QTLVRR457 pKa = 11.84NFSDD461 pKa = 3.49RR462 pKa = 11.84VSLLVDD468 pKa = 4.62LNSMIKK474 pKa = 8.78TVPPAGPYY482 pKa = 10.02PYY484 pKa = 9.67IQNAFLEE491 pKa = 4.7HH492 pKa = 6.94PFDD495 pKa = 4.22EE496 pKa = 4.59NAIGVDD502 pKa = 3.67EE503 pKa = 4.51YY504 pKa = 11.58DD505 pKa = 4.52GFVQDD510 pKa = 6.08DD511 pKa = 4.15VLCRR515 pKa = 11.84LPFQLDD521 pKa = 4.15PITTEE526 pKa = 3.9FKK528 pKa = 10.77KK529 pKa = 9.99MRR531 pKa = 11.84FAIEE535 pKa = 4.05IYY537 pKa = 10.59NPSTGVKK544 pKa = 10.02KK545 pKa = 10.4ILEE548 pKa = 4.33KK549 pKa = 10.65FDD551 pKa = 4.09VDD553 pKa = 3.37MTQFFDD559 pKa = 4.16DD560 pKa = 4.43ANGVPQFNLNQTRR573 pKa = 11.84GFKK576 pKa = 10.74LEE578 pKa = 4.08NGNNKK583 pKa = 8.9NWLKK587 pKa = 10.68INRR590 pKa = 11.84EE591 pKa = 3.78PSLDD595 pKa = 3.32SGGRR599 pKa = 11.84FGYY602 pKa = 10.45LAFYY606 pKa = 10.2AFKK609 pKa = 10.33IRR611 pKa = 11.84YY612 pKa = 7.6EE613 pKa = 3.89DD614 pKa = 3.82WINFPGMPNDD624 pKa = 4.01FFSSTEE630 pKa = 3.94LNNGFHH636 pKa = 7.07NDD638 pKa = 3.17WIHH641 pKa = 5.74YY642 pKa = 8.4FNTIGWQMNFYY653 pKa = 10.21TEE655 pKa = 3.67IDD657 pKa = 3.18AVEE660 pKa = 4.01NNEE663 pKa = 4.06LKK665 pKa = 10.43RR666 pKa = 11.84YY667 pKa = 8.9KK668 pKa = 10.77NKK670 pKa = 10.28FKK672 pKa = 11.0FKK674 pKa = 10.83FKK676 pKa = 10.74DD677 pKa = 3.2YY678 pKa = 11.24DD679 pKa = 3.98LNQLISTQHH688 pKa = 6.02RR689 pKa = 11.84YY690 pKa = 9.62YY691 pKa = 10.59RR692 pKa = 11.84DD693 pKa = 3.23SDD695 pKa = 3.67NTLLNVGTDD704 pKa = 4.42PISGRR709 pKa = 11.84PLGVIISNEE718 pKa = 3.8PTRR721 pKa = 11.84LEE723 pKa = 3.71IEE725 pKa = 4.23YY726 pKa = 10.26EE727 pKa = 4.06IMDD730 pKa = 4.12GGTWDD735 pKa = 3.24IANVYY740 pKa = 8.23ATATIEE746 pKa = 3.86IDD748 pKa = 3.01KK749 pKa = 10.85GAGIMQMRR757 pKa = 11.84QLSSVWGSEE766 pKa = 3.48NDD768 pKa = 3.41NPLIPLPGQTKK779 pKa = 10.54LKK781 pKa = 10.36IEE783 pKa = 4.0VDD785 pKa = 3.32ATLKK789 pKa = 10.9RR790 pKa = 11.84LTTKK794 pKa = 10.69CLIDD798 pKa = 5.02PDD800 pKa = 4.58LLDD803 pKa = 3.73DD804 pKa = 5.42AIRR807 pKa = 11.84YY808 pKa = 8.21RR809 pKa = 11.84ITGRR813 pKa = 11.84VGCFANGGSFEE824 pKa = 4.1FGKK827 pKa = 11.04YY828 pKa = 6.16EE829 pKa = 4.06ARR831 pKa = 11.84YY832 pKa = 7.89EE833 pKa = 4.16DD834 pKa = 4.57KK835 pKa = 11.59YY836 pKa = 11.13EE837 pKa = 3.81
MM1 pKa = 7.12VKK3 pKa = 10.44VIEE6 pKa = 4.15RR7 pKa = 11.84TYY9 pKa = 11.5NEE11 pKa = 3.89IFTPEE16 pKa = 3.98STSWLLGNVGEE27 pKa = 4.19WQKK30 pKa = 11.54LKK32 pKa = 11.1LKK34 pKa = 10.9VEE36 pKa = 4.22VSVDD40 pKa = 3.87LYY42 pKa = 11.17CDD44 pKa = 3.07QEE46 pKa = 4.01NRR48 pKa = 11.84LKK50 pKa = 10.18IDD52 pKa = 3.65YY53 pKa = 10.12INNSFVLNDD62 pKa = 3.51GKK64 pKa = 10.87KK65 pKa = 8.31WSDD68 pKa = 3.23YY69 pKa = 11.43GFDD72 pKa = 3.18IGQNIQLVYY81 pKa = 10.64KK82 pKa = 10.43FKK84 pKa = 11.14LDD86 pKa = 3.63TNGDD90 pKa = 4.01GIYY93 pKa = 10.22EE94 pKa = 4.37GPFYY98 pKa = 10.2QAPNFTIVNIASNVMEE114 pKa = 4.24VLQPIVIWVLADD126 pKa = 4.1TIPYY130 pKa = 8.93NQDD133 pKa = 2.34SVRR136 pKa = 11.84IIDD139 pKa = 3.77CKK141 pKa = 10.69FIAFVEE147 pKa = 4.36PEE149 pKa = 4.07GCRR152 pKa = 11.84IVYY155 pKa = 8.92GQISNEE161 pKa = 3.71NSEE164 pKa = 4.3NLSLQSFIDD173 pKa = 3.62GTTTMFVQPNVKK185 pKa = 9.93SQSLNINSSMNAVGMQSGMSVRR207 pKa = 11.84MVSVKK212 pKa = 10.44RR213 pKa = 11.84IGLKK217 pKa = 10.16PGSQNVYY224 pKa = 9.29QYY226 pKa = 11.08EE227 pKa = 4.12FEE229 pKa = 4.18LQYY232 pKa = 10.96LISSFFEE239 pKa = 4.43NISNFTNNQMPDD251 pKa = 3.37YY252 pKa = 10.39LLGDD256 pKa = 4.07GSLTDD261 pKa = 3.45NFQILFYY268 pKa = 10.36PKK270 pKa = 9.29WNNPNTFIQNDD281 pKa = 3.62MKK283 pKa = 10.43KK284 pKa = 8.0TARR287 pKa = 11.84LGNTGWFDD295 pKa = 3.56EE296 pKa = 4.86NYY298 pKa = 10.94NEE300 pKa = 4.82LEE302 pKa = 4.08NNFVIEE308 pKa = 4.06SLQYY312 pKa = 10.81FDD314 pKa = 6.38DD315 pKa = 4.01NGNPVDD321 pKa = 5.06AIDD324 pKa = 3.74YY325 pKa = 7.49FAPTKK330 pKa = 10.82VKK332 pKa = 10.74GIVSGVQNINTNTEE346 pKa = 3.97CGFGFIWVPKK356 pKa = 10.4FEE358 pKa = 4.5EE359 pKa = 4.73DD360 pKa = 3.91YY361 pKa = 11.21KK362 pKa = 11.42NKK364 pKa = 7.82EE365 pKa = 3.98TPFYY369 pKa = 11.18QNTFVNTGTTTDD381 pKa = 3.39GFKK384 pKa = 10.91VGTFYY389 pKa = 11.07PLTNVGGGLNSASIDD404 pKa = 3.57TKK406 pKa = 10.55NVKK409 pKa = 8.64FTDD412 pKa = 3.03IGGGKK417 pKa = 9.35ISFEE421 pKa = 4.52MIFTPNPNFYY431 pKa = 10.93LFFDD435 pKa = 4.38ARR437 pKa = 11.84NDD439 pKa = 3.71DD440 pKa = 3.53DD441 pKa = 4.51RR442 pKa = 11.84KK443 pKa = 9.43YY444 pKa = 11.16VLWLSVADD452 pKa = 3.6QTLVRR457 pKa = 11.84NFSDD461 pKa = 3.49RR462 pKa = 11.84VSLLVDD468 pKa = 4.62LNSMIKK474 pKa = 8.78TVPPAGPYY482 pKa = 10.02PYY484 pKa = 9.67IQNAFLEE491 pKa = 4.7HH492 pKa = 6.94PFDD495 pKa = 4.22EE496 pKa = 4.59NAIGVDD502 pKa = 3.67EE503 pKa = 4.51YY504 pKa = 11.58DD505 pKa = 4.52GFVQDD510 pKa = 6.08DD511 pKa = 4.15VLCRR515 pKa = 11.84LPFQLDD521 pKa = 4.15PITTEE526 pKa = 3.9FKK528 pKa = 10.77KK529 pKa = 9.99MRR531 pKa = 11.84FAIEE535 pKa = 4.05IYY537 pKa = 10.59NPSTGVKK544 pKa = 10.02KK545 pKa = 10.4ILEE548 pKa = 4.33KK549 pKa = 10.65FDD551 pKa = 4.09VDD553 pKa = 3.37MTQFFDD559 pKa = 4.16DD560 pKa = 4.43ANGVPQFNLNQTRR573 pKa = 11.84GFKK576 pKa = 10.74LEE578 pKa = 4.08NGNNKK583 pKa = 8.9NWLKK587 pKa = 10.68INRR590 pKa = 11.84EE591 pKa = 3.78PSLDD595 pKa = 3.32SGGRR599 pKa = 11.84FGYY602 pKa = 10.45LAFYY606 pKa = 10.2AFKK609 pKa = 10.33IRR611 pKa = 11.84YY612 pKa = 7.6EE613 pKa = 3.89DD614 pKa = 3.82WINFPGMPNDD624 pKa = 4.01FFSSTEE630 pKa = 3.94LNNGFHH636 pKa = 7.07NDD638 pKa = 3.17WIHH641 pKa = 5.74YY642 pKa = 8.4FNTIGWQMNFYY653 pKa = 10.21TEE655 pKa = 3.67IDD657 pKa = 3.18AVEE660 pKa = 4.01NNEE663 pKa = 4.06LKK665 pKa = 10.43RR666 pKa = 11.84YY667 pKa = 8.9KK668 pKa = 10.77NKK670 pKa = 10.28FKK672 pKa = 11.0FKK674 pKa = 10.83FKK676 pKa = 10.74DD677 pKa = 3.2YY678 pKa = 11.24DD679 pKa = 3.98LNQLISTQHH688 pKa = 6.02RR689 pKa = 11.84YY690 pKa = 9.62YY691 pKa = 10.59RR692 pKa = 11.84DD693 pKa = 3.23SDD695 pKa = 3.67NTLLNVGTDD704 pKa = 4.42PISGRR709 pKa = 11.84PLGVIISNEE718 pKa = 3.8PTRR721 pKa = 11.84LEE723 pKa = 3.71IEE725 pKa = 4.23YY726 pKa = 10.26EE727 pKa = 4.06IMDD730 pKa = 4.12GGTWDD735 pKa = 3.24IANVYY740 pKa = 8.23ATATIEE746 pKa = 3.86IDD748 pKa = 3.01KK749 pKa = 10.85GAGIMQMRR757 pKa = 11.84QLSSVWGSEE766 pKa = 3.48NDD768 pKa = 3.41NPLIPLPGQTKK779 pKa = 10.54LKK781 pKa = 10.36IEE783 pKa = 4.0VDD785 pKa = 3.32ATLKK789 pKa = 10.9RR790 pKa = 11.84LTTKK794 pKa = 10.69CLIDD798 pKa = 5.02PDD800 pKa = 4.58LLDD803 pKa = 3.73DD804 pKa = 5.42AIRR807 pKa = 11.84YY808 pKa = 8.21RR809 pKa = 11.84ITGRR813 pKa = 11.84VGCFANGGSFEE824 pKa = 4.1FGKK827 pKa = 11.04YY828 pKa = 6.16EE829 pKa = 4.06ARR831 pKa = 11.84YY832 pKa = 7.89EE833 pKa = 4.16DD834 pKa = 4.57KK835 pKa = 11.59YY836 pKa = 11.13EE837 pKa = 3.81
Molecular weight: 96.04 kDa
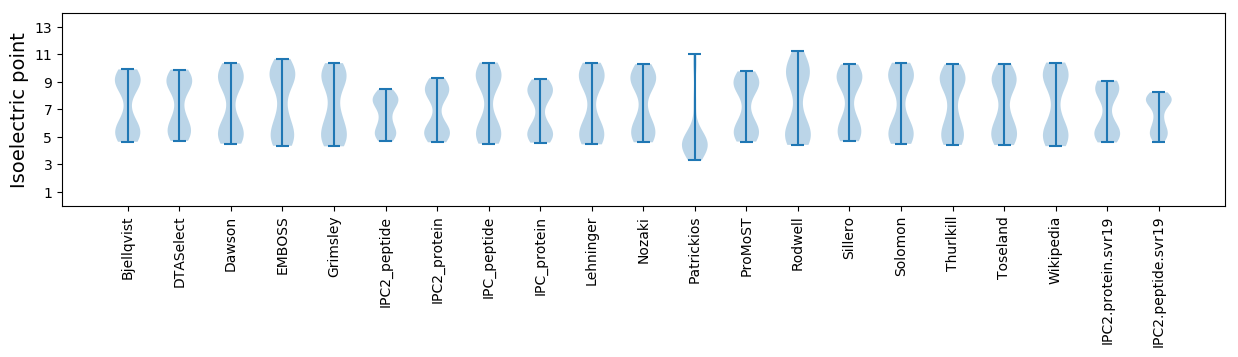
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B0WLH7|A0A1B0WLH7_9CAUD Uncharacterized protein OS=Flavobacterium phage Fpv6 OX=1814286 PE=4 SV=1
MM1 pKa = 7.58EE2 pKa = 5.63NYY4 pKa = 10.18NPSSQAKK11 pKa = 8.79LIRR14 pKa = 11.84MIFNKK19 pKa = 8.19NHH21 pKa = 6.76KK22 pKa = 9.85IILSDD27 pKa = 3.78EE28 pKa = 3.96QLKK31 pKa = 10.01TIDD34 pKa = 3.54IAFWDD39 pKa = 3.87YY40 pKa = 11.4KK41 pKa = 10.72KK42 pKa = 11.08EE43 pKa = 3.78KK44 pKa = 10.26LYY46 pKa = 10.89DD47 pKa = 2.99IGIYY51 pKa = 10.03RR52 pKa = 11.84GINICLFFILSLISFFTTTSKK73 pKa = 9.62WLKK76 pKa = 9.11IVLIIFFFYY85 pKa = 10.96NIIHH89 pKa = 7.09LLLTIKK95 pKa = 10.37RR96 pKa = 11.84YY97 pKa = 9.46KK98 pKa = 9.05QLKK101 pKa = 9.9NSFEE105 pKa = 4.09
MM1 pKa = 7.58EE2 pKa = 5.63NYY4 pKa = 10.18NPSSQAKK11 pKa = 8.79LIRR14 pKa = 11.84MIFNKK19 pKa = 8.19NHH21 pKa = 6.76KK22 pKa = 9.85IILSDD27 pKa = 3.78EE28 pKa = 3.96QLKK31 pKa = 10.01TIDD34 pKa = 3.54IAFWDD39 pKa = 3.87YY40 pKa = 11.4KK41 pKa = 10.72KK42 pKa = 11.08EE43 pKa = 3.78KK44 pKa = 10.26LYY46 pKa = 10.89DD47 pKa = 2.99IGIYY51 pKa = 10.03RR52 pKa = 11.84GINICLFFILSLISFFTTTSKK73 pKa = 9.62WLKK76 pKa = 9.11IVLIIFFFYY85 pKa = 10.96NIIHH89 pKa = 7.09LLLTIKK95 pKa = 10.37RR96 pKa = 11.84YY97 pKa = 9.46KK98 pKa = 9.05QLKK101 pKa = 9.9NSFEE105 pKa = 4.09
Molecular weight: 12.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13535 |
71 |
1103 |
233.4 |
26.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.625 ± 0.285 | 1.212 ± 0.162 |
5.807 ± 0.207 | 7.824 ± 0.417 |
5.696 ± 0.283 | 4.898 ± 0.325 |
1.322 ± 0.135 | 8.615 ± 0.272 |
9.945 ± 0.544 | 8.053 ± 0.309 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.268 ± 0.148 | 7.536 ± 0.363 |
2.985 ± 0.236 | 4.463 ± 0.385 |
3.258 ± 0.218 | 5.445 ± 0.228 |
5.911 ± 0.25 | 5.297 ± 0.253 |
1.093 ± 0.114 | 3.746 ± 0.214 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
