
Phaeodactylum tricornutum (strain CCAP 1055/1)
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Ochrophyta; Bacillariophyta; Bacillariophyceae; Bacillariophycidae; Naviculales; Phaeodactylaceae; Phaeodactylum; Phaeodactylum tricornutum
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
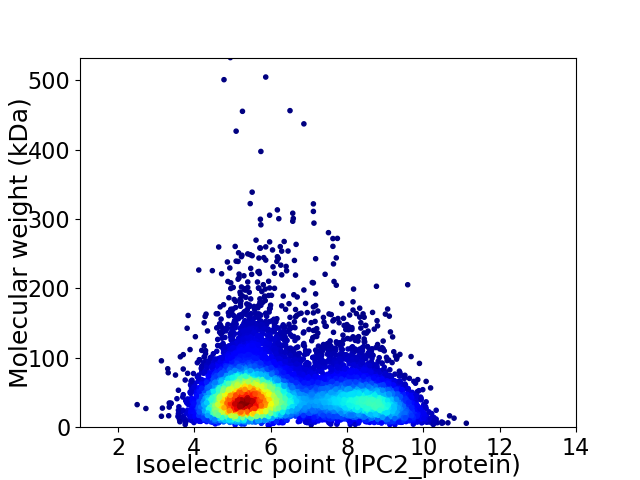
Virtual 2D-PAGE plot for 10465 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B7GBD6|B7GBD6_PHATC Predicted protein OS=Phaeodactylum tricornutum (strain CCAP 1055/1) OX=556484 GN=PHATRDRAFT_49665 PE=4 SV=1
MM1 pKa = 7.69RR2 pKa = 11.84GIFFAASTLVLGIASFRR19 pKa = 11.84ATDD22 pKa = 3.68AQDD25 pKa = 3.46VSCQLVRR32 pKa = 11.84QPNGSVEE39 pKa = 4.26YY40 pKa = 10.65VCDD43 pKa = 3.64QVRR46 pKa = 11.84EE47 pKa = 4.16PMRR50 pKa = 11.84PPLVPDD56 pKa = 3.97GQTYY60 pKa = 8.31QQGPVEE66 pKa = 4.96KK67 pKa = 10.56YY68 pKa = 10.28YY69 pKa = 10.75LQQPAEE75 pKa = 4.46DD76 pKa = 4.04VNASANATIPILRR89 pKa = 11.84RR90 pKa = 11.84VGVPPFDD97 pKa = 4.31TPLQACQGSCGSDD110 pKa = 2.97ADD112 pKa = 4.17CATGLVCLDD121 pKa = 3.27QLDD124 pKa = 4.01RR125 pKa = 11.84PRR127 pKa = 11.84DD128 pKa = 3.65GSVQGCSGNGRR139 pKa = 11.84LTMSVCVVPGSEE151 pKa = 3.92VDD153 pKa = 3.84LKK155 pKa = 10.86IGSLSLEE162 pKa = 4.35KK163 pKa = 11.01CQGTCSSDD171 pKa = 3.42DD172 pKa = 3.57DD173 pKa = 4.38CAGGLACFRR182 pKa = 11.84RR183 pKa = 11.84QGTEE187 pKa = 3.88TVPGCLEE194 pKa = 4.12RR195 pKa = 11.84DD196 pKa = 3.33VSEE199 pKa = 5.91SNFCHH204 pKa = 7.27DD205 pKa = 4.6PNEE208 pKa = 4.27TLARR212 pKa = 11.84LAFVGAAPHH221 pKa = 5.93SQSSLLPTCSGSCSSDD237 pKa = 2.71WDD239 pKa = 3.73CVRR242 pKa = 11.84GSNCFRR248 pKa = 11.84RR249 pKa = 11.84DD250 pKa = 3.12GTEE253 pKa = 3.89SVPGCEE259 pKa = 4.41GRR261 pKa = 11.84GLSGANYY268 pKa = 10.06CYY270 pKa = 10.1IAPEE274 pKa = 4.42GSLLLVGDD282 pKa = 4.43GVTYY286 pKa = 10.19IQYY289 pKa = 10.28PLRR292 pKa = 11.84QCEE295 pKa = 4.11GHH297 pKa = 6.96CIADD301 pKa = 3.66IDD303 pKa = 4.37CEE305 pKa = 4.35SGLKK309 pKa = 10.11CFRR312 pKa = 11.84RR313 pKa = 11.84EE314 pKa = 4.06GSEE317 pKa = 4.73HH318 pKa = 7.04IPGCDD323 pKa = 3.19GEE325 pKa = 4.72GADD328 pKa = 4.13RR329 pKa = 11.84TNYY332 pKa = 9.89CYY334 pKa = 10.36QPFPDD339 pKa = 3.88TSGPTDD345 pKa = 4.05GPTLAPLSVSPSNSPSLILTLDD367 pKa = 3.54PTNSLGSRR375 pKa = 11.84GGEE378 pKa = 3.94SEE380 pKa = 4.59SPSTPTSDD388 pKa = 3.5MPSISPSDD396 pKa = 4.22EE397 pKa = 3.71PSMIPSDD404 pKa = 4.16TPSMVPSDD412 pKa = 4.5VPSMLPSDD420 pKa = 4.74APSMIPSDD428 pKa = 4.36SPSDD432 pKa = 3.75TPSDD436 pKa = 3.8VPSDD440 pKa = 3.99VPSVLPSFSPSMRR453 pKa = 11.84PSDD456 pKa = 4.09TPSTMPSDD464 pKa = 3.86TPTDD468 pKa = 3.81VPSDD472 pKa = 3.94APSDD476 pKa = 4.03SPSDD480 pKa = 3.75VPSDD484 pKa = 3.95VPSDD488 pKa = 3.82APSDD492 pKa = 3.99VPSDD496 pKa = 3.99VPSDD500 pKa = 3.82APSDD504 pKa = 4.09VPSDD508 pKa = 3.81TPSDD512 pKa = 3.91VPSDD516 pKa = 4.05VPSDD520 pKa = 3.81TPSDD524 pKa = 3.91VPSDD528 pKa = 4.05VPSDD532 pKa = 3.71TPSAVPSDD540 pKa = 4.06VPSGNPSDD548 pKa = 4.46VPSEE552 pKa = 4.33TPSDD556 pKa = 4.04VPSDD560 pKa = 3.88TPSDD564 pKa = 3.91VPSDD568 pKa = 3.92VPSDD572 pKa = 3.87VPSDD576 pKa = 3.87VPSDD580 pKa = 3.87VPSDD584 pKa = 3.87VPSDD588 pKa = 3.96VPSDD592 pKa = 3.81TPSDD596 pKa = 3.91VPSDD600 pKa = 4.05VPSDD604 pKa = 3.87TPSDD608 pKa = 3.94LPSDD612 pKa = 4.16VPSDD616 pKa = 3.97VPSTMPSSTLGGTSSNGPVTGKK638 pKa = 10.9GSLAPTGSFSGEE650 pKa = 3.82SSGRR654 pKa = 11.84PSKK657 pKa = 10.09TPGVMVEE664 pKa = 3.9LSARR668 pKa = 11.84PSEE671 pKa = 4.49SFTPSPTTFPVLLPSKK687 pKa = 9.82VPSIAPSRR695 pKa = 11.84TEE697 pKa = 3.74TVAPSTAFPTEE708 pKa = 4.27TSSSLPTLTTTVSSAPTQMCSMSASEE734 pKa = 4.08RR735 pKa = 11.84ASTIMGILEE744 pKa = 3.99EE745 pKa = 4.82DD746 pKa = 4.08FNDD749 pKa = 3.29VSSPQFRR756 pKa = 11.84AVEE759 pKa = 4.02WLVQIDD765 pKa = 4.44PLSLCPGDD773 pKa = 4.27EE774 pKa = 4.27NLEE777 pKa = 3.78QRR779 pKa = 11.84YY780 pKa = 9.1ILAVLYY786 pKa = 8.81FQTGGEE792 pKa = 3.61WWTRR796 pKa = 11.84CSPMSAEE803 pKa = 4.09VCDD806 pKa = 3.56QGEE809 pKa = 4.29AFLSGANEE817 pKa = 4.29CAWGGVNCDD826 pKa = 2.79SSSRR830 pKa = 11.84VTALHH835 pKa = 6.83LDD837 pKa = 3.85SNNLSGSLPSEE848 pKa = 4.62LGRR851 pKa = 11.84LAYY854 pKa = 10.24LVEE857 pKa = 5.8LDD859 pKa = 3.84MDD861 pKa = 4.99DD862 pKa = 4.97NEE864 pKa = 4.68LTGSIPRR871 pKa = 11.84ILGQLSFLEE880 pKa = 5.02IVDD883 pKa = 5.28LDD885 pKa = 4.26DD886 pKa = 4.08NQLTGSIPEE895 pKa = 4.0EE896 pKa = 4.32LYY898 pKa = 10.85SVSSLEE904 pKa = 5.28ILDD907 pKa = 5.23LDD909 pKa = 3.96INQLTGTISTLIGNLVNLYY928 pKa = 10.01YY929 pKa = 10.86LQIDD933 pKa = 3.92SNKK936 pKa = 8.07FTGSIPSEE944 pKa = 4.14VGTLTRR950 pKa = 11.84LEE952 pKa = 4.39YY953 pKa = 10.77FSMTDD958 pKa = 3.07IQIAEE963 pKa = 4.51ALPDD967 pKa = 3.91SLCSRR972 pKa = 11.84DD973 pKa = 3.58TLLLFGDD980 pKa = 4.5CEE982 pKa = 4.4VCVVEE987 pKa = 5.33DD988 pKa = 4.06CCTACLAKK996 pKa = 9.73EE997 pKa = 4.25TTPP1000 pKa = 4.07
MM1 pKa = 7.69RR2 pKa = 11.84GIFFAASTLVLGIASFRR19 pKa = 11.84ATDD22 pKa = 3.68AQDD25 pKa = 3.46VSCQLVRR32 pKa = 11.84QPNGSVEE39 pKa = 4.26YY40 pKa = 10.65VCDD43 pKa = 3.64QVRR46 pKa = 11.84EE47 pKa = 4.16PMRR50 pKa = 11.84PPLVPDD56 pKa = 3.97GQTYY60 pKa = 8.31QQGPVEE66 pKa = 4.96KK67 pKa = 10.56YY68 pKa = 10.28YY69 pKa = 10.75LQQPAEE75 pKa = 4.46DD76 pKa = 4.04VNASANATIPILRR89 pKa = 11.84RR90 pKa = 11.84VGVPPFDD97 pKa = 4.31TPLQACQGSCGSDD110 pKa = 2.97ADD112 pKa = 4.17CATGLVCLDD121 pKa = 3.27QLDD124 pKa = 4.01RR125 pKa = 11.84PRR127 pKa = 11.84DD128 pKa = 3.65GSVQGCSGNGRR139 pKa = 11.84LTMSVCVVPGSEE151 pKa = 3.92VDD153 pKa = 3.84LKK155 pKa = 10.86IGSLSLEE162 pKa = 4.35KK163 pKa = 11.01CQGTCSSDD171 pKa = 3.42DD172 pKa = 3.57DD173 pKa = 4.38CAGGLACFRR182 pKa = 11.84RR183 pKa = 11.84QGTEE187 pKa = 3.88TVPGCLEE194 pKa = 4.12RR195 pKa = 11.84DD196 pKa = 3.33VSEE199 pKa = 5.91SNFCHH204 pKa = 7.27DD205 pKa = 4.6PNEE208 pKa = 4.27TLARR212 pKa = 11.84LAFVGAAPHH221 pKa = 5.93SQSSLLPTCSGSCSSDD237 pKa = 2.71WDD239 pKa = 3.73CVRR242 pKa = 11.84GSNCFRR248 pKa = 11.84RR249 pKa = 11.84DD250 pKa = 3.12GTEE253 pKa = 3.89SVPGCEE259 pKa = 4.41GRR261 pKa = 11.84GLSGANYY268 pKa = 10.06CYY270 pKa = 10.1IAPEE274 pKa = 4.42GSLLLVGDD282 pKa = 4.43GVTYY286 pKa = 10.19IQYY289 pKa = 10.28PLRR292 pKa = 11.84QCEE295 pKa = 4.11GHH297 pKa = 6.96CIADD301 pKa = 3.66IDD303 pKa = 4.37CEE305 pKa = 4.35SGLKK309 pKa = 10.11CFRR312 pKa = 11.84RR313 pKa = 11.84EE314 pKa = 4.06GSEE317 pKa = 4.73HH318 pKa = 7.04IPGCDD323 pKa = 3.19GEE325 pKa = 4.72GADD328 pKa = 4.13RR329 pKa = 11.84TNYY332 pKa = 9.89CYY334 pKa = 10.36QPFPDD339 pKa = 3.88TSGPTDD345 pKa = 4.05GPTLAPLSVSPSNSPSLILTLDD367 pKa = 3.54PTNSLGSRR375 pKa = 11.84GGEE378 pKa = 3.94SEE380 pKa = 4.59SPSTPTSDD388 pKa = 3.5MPSISPSDD396 pKa = 4.22EE397 pKa = 3.71PSMIPSDD404 pKa = 4.16TPSMVPSDD412 pKa = 4.5VPSMLPSDD420 pKa = 4.74APSMIPSDD428 pKa = 4.36SPSDD432 pKa = 3.75TPSDD436 pKa = 3.8VPSDD440 pKa = 3.99VPSVLPSFSPSMRR453 pKa = 11.84PSDD456 pKa = 4.09TPSTMPSDD464 pKa = 3.86TPTDD468 pKa = 3.81VPSDD472 pKa = 3.94APSDD476 pKa = 4.03SPSDD480 pKa = 3.75VPSDD484 pKa = 3.95VPSDD488 pKa = 3.82APSDD492 pKa = 3.99VPSDD496 pKa = 3.99VPSDD500 pKa = 3.82APSDD504 pKa = 4.09VPSDD508 pKa = 3.81TPSDD512 pKa = 3.91VPSDD516 pKa = 4.05VPSDD520 pKa = 3.81TPSDD524 pKa = 3.91VPSDD528 pKa = 4.05VPSDD532 pKa = 3.71TPSAVPSDD540 pKa = 4.06VPSGNPSDD548 pKa = 4.46VPSEE552 pKa = 4.33TPSDD556 pKa = 4.04VPSDD560 pKa = 3.88TPSDD564 pKa = 3.91VPSDD568 pKa = 3.92VPSDD572 pKa = 3.87VPSDD576 pKa = 3.87VPSDD580 pKa = 3.87VPSDD584 pKa = 3.87VPSDD588 pKa = 3.96VPSDD592 pKa = 3.81TPSDD596 pKa = 3.91VPSDD600 pKa = 4.05VPSDD604 pKa = 3.87TPSDD608 pKa = 3.94LPSDD612 pKa = 4.16VPSDD616 pKa = 3.97VPSTMPSSTLGGTSSNGPVTGKK638 pKa = 10.9GSLAPTGSFSGEE650 pKa = 3.82SSGRR654 pKa = 11.84PSKK657 pKa = 10.09TPGVMVEE664 pKa = 3.9LSARR668 pKa = 11.84PSEE671 pKa = 4.49SFTPSPTTFPVLLPSKK687 pKa = 9.82VPSIAPSRR695 pKa = 11.84TEE697 pKa = 3.74TVAPSTAFPTEE708 pKa = 4.27TSSSLPTLTTTVSSAPTQMCSMSASEE734 pKa = 4.08RR735 pKa = 11.84ASTIMGILEE744 pKa = 3.99EE745 pKa = 4.82DD746 pKa = 4.08FNDD749 pKa = 3.29VSSPQFRR756 pKa = 11.84AVEE759 pKa = 4.02WLVQIDD765 pKa = 4.44PLSLCPGDD773 pKa = 4.27EE774 pKa = 4.27NLEE777 pKa = 3.78QRR779 pKa = 11.84YY780 pKa = 9.1ILAVLYY786 pKa = 8.81FQTGGEE792 pKa = 3.61WWTRR796 pKa = 11.84CSPMSAEE803 pKa = 4.09VCDD806 pKa = 3.56QGEE809 pKa = 4.29AFLSGANEE817 pKa = 4.29CAWGGVNCDD826 pKa = 2.79SSSRR830 pKa = 11.84VTALHH835 pKa = 6.83LDD837 pKa = 3.85SNNLSGSLPSEE848 pKa = 4.62LGRR851 pKa = 11.84LAYY854 pKa = 10.24LVEE857 pKa = 5.8LDD859 pKa = 3.84MDD861 pKa = 4.99DD862 pKa = 4.97NEE864 pKa = 4.68LTGSIPRR871 pKa = 11.84ILGQLSFLEE880 pKa = 5.02IVDD883 pKa = 5.28LDD885 pKa = 4.26DD886 pKa = 4.08NQLTGSIPEE895 pKa = 4.0EE896 pKa = 4.32LYY898 pKa = 10.85SVSSLEE904 pKa = 5.28ILDD907 pKa = 5.23LDD909 pKa = 3.96INQLTGTISTLIGNLVNLYY928 pKa = 10.01YY929 pKa = 10.86LQIDD933 pKa = 3.92SNKK936 pKa = 8.07FTGSIPSEE944 pKa = 4.14VGTLTRR950 pKa = 11.84LEE952 pKa = 4.39YY953 pKa = 10.77FSMTDD958 pKa = 3.07IQIAEE963 pKa = 4.51ALPDD967 pKa = 3.91SLCSRR972 pKa = 11.84DD973 pKa = 3.58TLLLFGDD980 pKa = 4.5CEE982 pKa = 4.4VCVVEE987 pKa = 5.33DD988 pKa = 4.06CCTACLAKK996 pKa = 9.73EE997 pKa = 4.25TTPP1000 pKa = 4.07
Molecular weight: 104.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B5Y3A4|B5Y3A4_PHATC Predicted protein (Fragment) OS=Phaeodactylum tricornutum (strain CCAP 1055/1) OX=556484 GN=PHATR_13439 PE=4 SV=1
MM1 pKa = 7.5TKK3 pKa = 9.03RR4 pKa = 11.84TLSNKK9 pKa = 9.3SRR11 pKa = 11.84YY12 pKa = 9.39SVLKK16 pKa = 10.53LSGFRR21 pKa = 11.84SRR23 pKa = 11.84MATPQGRR30 pKa = 11.84KK31 pKa = 7.39TIRR34 pKa = 11.84NRR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.85KK39 pKa = 10.07GRR41 pKa = 11.84KK42 pKa = 8.84NLTLRR47 pKa = 11.84RR48 pKa = 3.92
MM1 pKa = 7.5TKK3 pKa = 9.03RR4 pKa = 11.84TLSNKK9 pKa = 9.3SRR11 pKa = 11.84YY12 pKa = 9.39SVLKK16 pKa = 10.53LSGFRR21 pKa = 11.84SRR23 pKa = 11.84MATPQGRR30 pKa = 11.84KK31 pKa = 7.39TIRR34 pKa = 11.84NRR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.85KK39 pKa = 10.07GRR41 pKa = 11.84KK42 pKa = 8.84NLTLRR47 pKa = 11.84RR48 pKa = 3.92
Molecular weight: 5.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4838762 |
29 |
4825 |
462.4 |
51.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.512 ± 0.02 | 1.569 ± 0.01 |
5.893 ± 0.016 | 6.083 ± 0.019 |
3.747 ± 0.013 | 6.394 ± 0.021 |
2.462 ± 0.011 | 4.53 ± 0.015 |
4.659 ± 0.018 | 9.452 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.253 ± 0.009 | 3.785 ± 0.012 |
5.188 ± 0.022 | 4.091 ± 0.014 |
6.148 ± 0.017 | 8.372 ± 0.026 |
6.125 ± 0.015 | 6.834 ± 0.017 |
1.347 ± 0.009 | 2.555 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
