
Auriculariopsis ampla
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Agaricales; Schizophyllaceae; Auriculariopsis
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
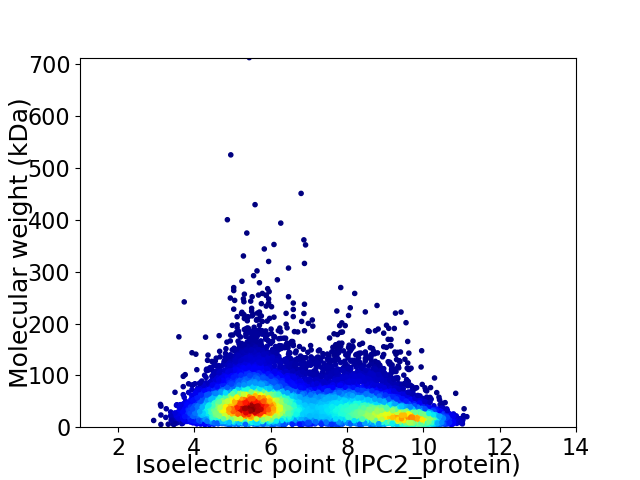
Virtual 2D-PAGE plot for 15404 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A550CAU4|A0A550CAU4_9AGAR Asparagine synthase (glutamine-hydrolyzing) OS=Auriculariopsis ampla OX=97359 GN=BD626DRAFT_500924 PE=4 SV=1
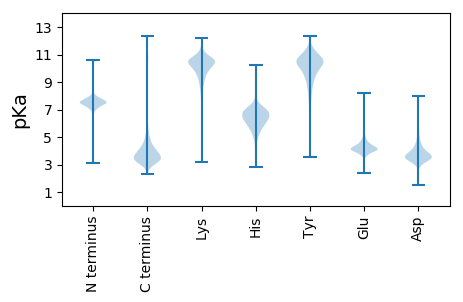
MM1 pKa = 7.59PSEE4 pKa = 4.24STNLFNLARR13 pKa = 11.84NKK15 pKa = 10.36LHH17 pKa = 6.98LSLGSRR23 pKa = 11.84SDD25 pKa = 3.69CSLHH29 pKa = 6.71RR30 pKa = 11.84WVLLKK35 pKa = 10.97NSIVHH40 pKa = 5.35STPHH44 pKa = 6.4SSSAPDD50 pKa = 3.42VAASDD55 pKa = 3.52AAADD59 pKa = 3.72VRR61 pKa = 11.84NAPDD65 pKa = 3.0TDD67 pKa = 3.81YY68 pKa = 11.1TVEE71 pKa = 4.09EE72 pKa = 4.37EE73 pKa = 4.12VDD75 pKa = 3.59SFMFPDD81 pKa = 4.52ADD83 pKa = 3.9QLIDD87 pKa = 4.01DD88 pKa = 5.13RR89 pKa = 11.84AAEE92 pKa = 4.38GNGSEE97 pKa = 4.29EE98 pKa = 3.86QWLDD102 pKa = 3.44SLLEE106 pKa = 4.2TLSHH110 pKa = 6.57EE111 pKa = 4.4EE112 pKa = 3.84EE113 pKa = 5.21DD114 pKa = 4.48DD115 pKa = 4.0FGMDD119 pKa = 3.54SDD121 pKa = 4.24VHH123 pKa = 8.03SMLAEE128 pKa = 5.02DD129 pKa = 5.84DD130 pKa = 5.35DD131 pKa = 6.2DD132 pKa = 6.74DD133 pKa = 5.66SLLSPSISPMSSSDD147 pKa = 3.59DD148 pKa = 3.46LTRR151 pKa = 11.84PPVYY155 pKa = 9.86YY156 pKa = 9.28PPPIAVPYY164 pKa = 7.79PVPYY168 pKa = 9.89PPYY171 pKa = 9.23QPPPARR177 pKa = 11.84ADD179 pKa = 3.58ALPSPFDD186 pKa = 3.25IHH188 pKa = 7.9RR189 pKa = 11.84MSPYY193 pKa = 10.52LDD195 pKa = 3.85PLPYY199 pKa = 9.94HH200 pKa = 6.88DD201 pKa = 6.14AEE203 pKa = 6.61DD204 pKa = 4.0EE205 pKa = 4.58DD206 pKa = 5.38EE207 pKa = 4.75DD208 pKa = 6.17LSMPDD213 pKa = 4.23AIEE216 pKa = 4.43DD217 pKa = 3.91TSDD220 pKa = 4.21DD221 pKa = 4.09EE222 pKa = 5.09SDD224 pKa = 3.86SPLTPSIGHH233 pKa = 6.17SGSLSQDD240 pKa = 3.48PAQVPLPGEE249 pKa = 3.73RR250 pKa = 11.84SGLRR254 pKa = 11.84EE255 pKa = 3.96HH256 pKa = 6.35VHH258 pKa = 7.2ILVDD262 pKa = 3.45TDD264 pKa = 4.0DD265 pKa = 3.99SYY267 pKa = 11.9FYY269 pKa = 10.58PFAVDD274 pKa = 4.29PLPFPDD280 pKa = 5.36DD281 pKa = 3.63QLQNPYY287 pKa = 8.53NTYY290 pKa = 9.51QEE292 pKa = 4.51CC293 pKa = 3.52
MM1 pKa = 7.59PSEE4 pKa = 4.24STNLFNLARR13 pKa = 11.84NKK15 pKa = 10.36LHH17 pKa = 6.98LSLGSRR23 pKa = 11.84SDD25 pKa = 3.69CSLHH29 pKa = 6.71RR30 pKa = 11.84WVLLKK35 pKa = 10.97NSIVHH40 pKa = 5.35STPHH44 pKa = 6.4SSSAPDD50 pKa = 3.42VAASDD55 pKa = 3.52AAADD59 pKa = 3.72VRR61 pKa = 11.84NAPDD65 pKa = 3.0TDD67 pKa = 3.81YY68 pKa = 11.1TVEE71 pKa = 4.09EE72 pKa = 4.37EE73 pKa = 4.12VDD75 pKa = 3.59SFMFPDD81 pKa = 4.52ADD83 pKa = 3.9QLIDD87 pKa = 4.01DD88 pKa = 5.13RR89 pKa = 11.84AAEE92 pKa = 4.38GNGSEE97 pKa = 4.29EE98 pKa = 3.86QWLDD102 pKa = 3.44SLLEE106 pKa = 4.2TLSHH110 pKa = 6.57EE111 pKa = 4.4EE112 pKa = 3.84EE113 pKa = 5.21DD114 pKa = 4.48DD115 pKa = 4.0FGMDD119 pKa = 3.54SDD121 pKa = 4.24VHH123 pKa = 8.03SMLAEE128 pKa = 5.02DD129 pKa = 5.84DD130 pKa = 5.35DD131 pKa = 6.2DD132 pKa = 6.74DD133 pKa = 5.66SLLSPSISPMSSSDD147 pKa = 3.59DD148 pKa = 3.46LTRR151 pKa = 11.84PPVYY155 pKa = 9.86YY156 pKa = 9.28PPPIAVPYY164 pKa = 7.79PVPYY168 pKa = 9.89PPYY171 pKa = 9.23QPPPARR177 pKa = 11.84ADD179 pKa = 3.58ALPSPFDD186 pKa = 3.25IHH188 pKa = 7.9RR189 pKa = 11.84MSPYY193 pKa = 10.52LDD195 pKa = 3.85PLPYY199 pKa = 9.94HH200 pKa = 6.88DD201 pKa = 6.14AEE203 pKa = 6.61DD204 pKa = 4.0EE205 pKa = 4.58DD206 pKa = 5.38EE207 pKa = 4.75DD208 pKa = 6.17LSMPDD213 pKa = 4.23AIEE216 pKa = 4.43DD217 pKa = 3.91TSDD220 pKa = 4.21DD221 pKa = 4.09EE222 pKa = 5.09SDD224 pKa = 3.86SPLTPSIGHH233 pKa = 6.17SGSLSQDD240 pKa = 3.48PAQVPLPGEE249 pKa = 3.73RR250 pKa = 11.84SGLRR254 pKa = 11.84EE255 pKa = 3.96HH256 pKa = 6.35VHH258 pKa = 7.2ILVDD262 pKa = 3.45TDD264 pKa = 4.0DD265 pKa = 3.99SYY267 pKa = 11.9FYY269 pKa = 10.58PFAVDD274 pKa = 4.29PLPFPDD280 pKa = 5.36DD281 pKa = 3.63QLQNPYY287 pKa = 8.53NTYY290 pKa = 9.51QEE292 pKa = 4.51CC293 pKa = 3.52
Molecular weight: 32.4 kDa
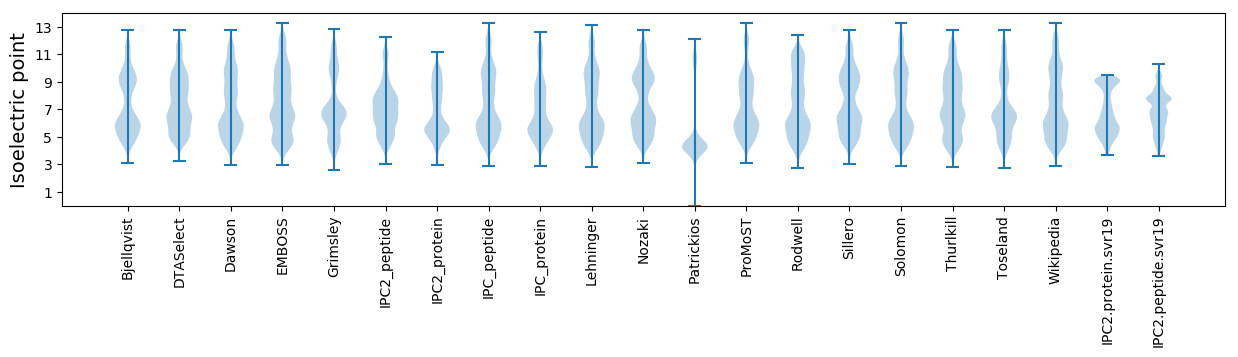
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A550C2I8|A0A550C2I8_9AGAR Protein kinase domain-containing protein OS=Auriculariopsis ampla OX=97359 GN=BD626DRAFT_573039 PE=4 SV=1
MM1 pKa = 7.08SQSTNLTRR9 pKa = 11.84PSSARR14 pKa = 11.84TTHH17 pKa = 6.93TIQLTTAASHH27 pKa = 6.25GAPRR31 pKa = 11.84MTASARR37 pKa = 11.84ALTGKK42 pKa = 10.12SSARR46 pKa = 11.84STPTTTIAASLSKK59 pKa = 10.47RR60 pKa = 11.84RR61 pKa = 11.84NN62 pKa = 3.24
MM1 pKa = 7.08SQSTNLTRR9 pKa = 11.84PSSARR14 pKa = 11.84TTHH17 pKa = 6.93TIQLTTAASHH27 pKa = 6.25GAPRR31 pKa = 11.84MTASARR37 pKa = 11.84ALTGKK42 pKa = 10.12SSARR46 pKa = 11.84STPTTTIAASLSKK59 pKa = 10.47RR60 pKa = 11.84RR61 pKa = 11.84NN62 pKa = 3.24
Molecular weight: 6.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6453724 |
49 |
6535 |
419.0 |
46.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.93 ± 0.021 | 1.42 ± 0.009 |
5.914 ± 0.016 | 5.736 ± 0.019 |
3.384 ± 0.013 | 6.519 ± 0.022 |
2.615 ± 0.01 | 4.203 ± 0.014 |
3.948 ± 0.018 | 9.038 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.256 ± 0.008 | 2.909 ± 0.009 |
6.777 ± 0.026 | 3.578 ± 0.012 |
7.093 ± 0.02 | 8.265 ± 0.027 |
5.944 ± 0.012 | 6.377 ± 0.015 |
1.429 ± 0.008 | 2.663 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
