
Eimeria maxima (Coccidian parasite)
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Conoidasida; Coccidia; Eucoccidiorida; Eimeriorina; Eimeriidae; Eimeria
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
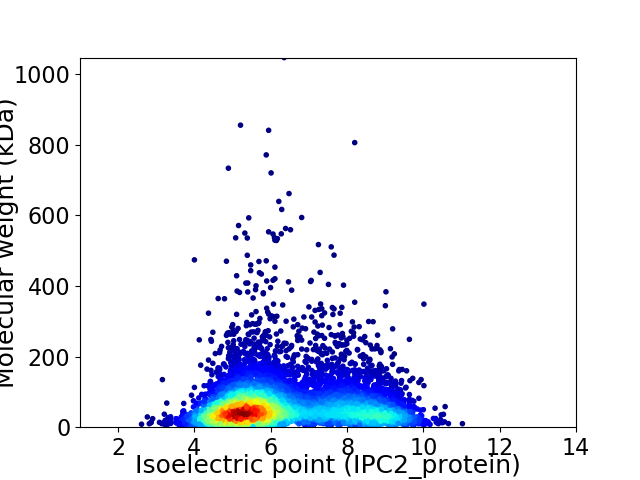
Virtual 2D-PAGE plot for 6057 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U6M8M7|U6M8M7_EIMMA Ectonucleoside triphosphate diphosphohydrolase putative OS=Eimeria maxima OX=5804 GN=EMWEY_00006300 PE=4 SV=1
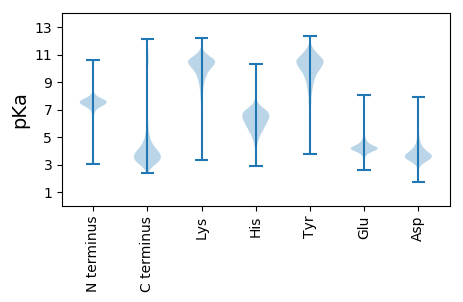
MM1 pKa = 7.88GDD3 pKa = 3.49GLEE6 pKa = 4.05QQEE9 pKa = 5.35DD10 pKa = 3.71KK11 pKa = 11.05QQQQQQQQQQQQQQQQQQQQQQQQQQQHH39 pKa = 6.91DD40 pKa = 4.07DD41 pKa = 3.68DD42 pKa = 6.24DD43 pKa = 4.42FVLDD47 pKa = 4.78LDD49 pKa = 5.04EE50 pKa = 6.59DD51 pKa = 3.87EE52 pKa = 4.95QDD54 pKa = 3.7NSKK57 pKa = 9.51QQQQQQQQQQQQQQQQEE74 pKa = 4.28QQQEE78 pKa = 4.25CSNNGSICVSDD89 pKa = 4.34AEE91 pKa = 4.47SSVAASAAPASSAATAAAAATAAAAAAWDD120 pKa = 4.04RR121 pKa = 11.84EE122 pKa = 4.32SQEE125 pKa = 4.05EE126 pKa = 4.51GLVDD130 pKa = 4.15MWNMLHH136 pKa = 7.48PDD138 pKa = 4.34GPQQQQQQQQQQQQQQQQQQQQQGSEE164 pKa = 4.43KK165 pKa = 10.47YY166 pKa = 9.33PLSPTDD172 pKa = 3.09TSYY175 pKa = 10.71TYY177 pKa = 10.17PHH179 pKa = 6.78HH180 pKa = 6.31QQQQQQQQQEE190 pKa = 4.28QQQQQQQHH198 pKa = 4.78QSHH201 pKa = 6.46NNDD204 pKa = 2.65IPFDD208 pKa = 3.94FFNINLFNPLPAAMVSNNN226 pKa = 3.11
MM1 pKa = 7.88GDD3 pKa = 3.49GLEE6 pKa = 4.05QQEE9 pKa = 5.35DD10 pKa = 3.71KK11 pKa = 11.05QQQQQQQQQQQQQQQQQQQQQQQQQQQHH39 pKa = 6.91DD40 pKa = 4.07DD41 pKa = 3.68DD42 pKa = 6.24DD43 pKa = 4.42FVLDD47 pKa = 4.78LDD49 pKa = 5.04EE50 pKa = 6.59DD51 pKa = 3.87EE52 pKa = 4.95QDD54 pKa = 3.7NSKK57 pKa = 9.51QQQQQQQQQQQQQQQQEE74 pKa = 4.28QQQEE78 pKa = 4.25CSNNGSICVSDD89 pKa = 4.34AEE91 pKa = 4.47SSVAASAAPASSAATAAAAATAAAAAAWDD120 pKa = 4.04RR121 pKa = 11.84EE122 pKa = 4.32SQEE125 pKa = 4.05EE126 pKa = 4.51GLVDD130 pKa = 4.15MWNMLHH136 pKa = 7.48PDD138 pKa = 4.34GPQQQQQQQQQQQQQQQQQQQQQGSEE164 pKa = 4.43KK165 pKa = 10.47YY166 pKa = 9.33PLSPTDD172 pKa = 3.09TSYY175 pKa = 10.71TYY177 pKa = 10.17PHH179 pKa = 6.78HH180 pKa = 6.31QQQQQQQQQEE190 pKa = 4.28QQQQQQQHH198 pKa = 4.78QSHH201 pKa = 6.46NNDD204 pKa = 2.65IPFDD208 pKa = 3.94FFNINLFNPLPAAMVSNNN226 pKa = 3.11
Molecular weight: 26.01 kDa
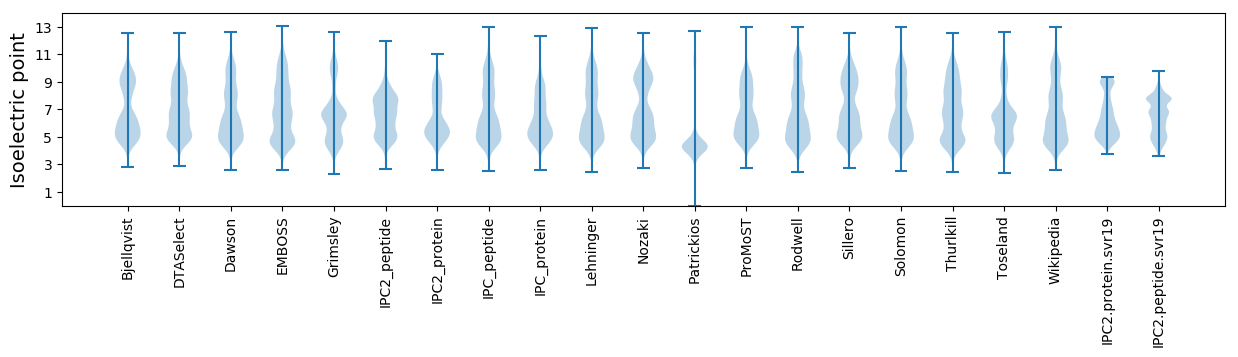
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U6M688|U6M688_EIMMA Uncharacterized protein OS=Eimeria maxima OX=5804 GN=EMWEY_00046210 PE=4 SV=1
MM1 pKa = 7.43SRR3 pKa = 11.84LGLSKK8 pKa = 10.9ALLRR12 pKa = 11.84FSLARR17 pKa = 11.84LRR19 pKa = 11.84PAATAAAAAATAATTAPAPSRR40 pKa = 11.84RR41 pKa = 11.84LLSSLSAAAPAAAAAGAAAAPAAAAGTEE69 pKa = 4.17TSPGVRR75 pKa = 11.84PPSLEE80 pKa = 3.74GKK82 pKa = 9.92FFTPAKK88 pKa = 9.09PWGGEE93 pKa = 3.77QQQQQQQQQHH103 pKa = 5.7LQQQQHH109 pKa = 5.59LHH111 pKa = 4.86QQQQHH116 pKa = 5.29LHH118 pKa = 4.74QQQQHH123 pKa = 5.27FHH125 pKa = 5.05QQQHH129 pKa = 5.48LHH131 pKa = 5.15QQQQLHH137 pKa = 5.12QQQQHH142 pKa = 5.13LHH144 pKa = 5.42HH145 pKa = 6.71HH146 pKa = 5.28QQQLHH151 pKa = 5.94HH152 pKa = 5.79QQQLHH157 pKa = 4.55QQQHH161 pKa = 5.23LHH163 pKa = 5.08QQQQLHH169 pKa = 6.12HH170 pKa = 5.63QQQLHH175 pKa = 4.81QQQQHH180 pKa = 5.34LHH182 pKa = 4.7QQQHH186 pKa = 5.41LHH188 pKa = 4.98QQQHH192 pKa = 5.81LQQQLQQQAQQQQQRR207 pKa = 11.84GRR209 pKa = 11.84GAFLPADD216 pKa = 4.31DD217 pKa = 4.56PWYY220 pKa = 9.76SSSYY224 pKa = 10.26EE225 pKa = 4.04PVSPPPRR232 pKa = 11.84QQQQQQQQQQQQQHH246 pKa = 5.16WRR248 pKa = 11.84PSAATAAAGGPGAGLRR264 pKa = 11.84PIDD267 pKa = 3.63WSKK270 pKa = 11.82KK271 pKa = 7.87NLPPIYY277 pKa = 9.33PVAPPPPPAAAAAAAAPGAAAAAAAAAATSAARR310 pKa = 11.84MKK312 pKa = 10.54EE313 pKa = 3.81LGIKK317 pKa = 9.91IEE319 pKa = 4.48GNAPLPLLLPASLPAAFAAAGLPDD343 pKa = 4.77LLLHH347 pKa = 7.04ALQHH351 pKa = 6.73LPQQQQQQQQQQQQQQQQISTLFPIQQIGWSCCFTGRR388 pKa = 11.84DD389 pKa = 4.19LIAIAQTGSGKK400 pKa = 9.22TIGYY404 pKa = 7.78LIPALIHH411 pKa = 6.94LLRR414 pKa = 11.84QQQQQQLQQQQQQQQGNSSSSSSSSSSRR442 pKa = 11.84GPTVLIVGPTRR453 pKa = 11.84EE454 pKa = 4.32LVQQIHH460 pKa = 6.5AEE462 pKa = 4.06CQRR465 pKa = 11.84ILKK468 pKa = 9.82AAAAAAAARR477 pKa = 11.84ASAAAAGGTATAAAAAAAAAAANTPLARR505 pKa = 11.84CVAVYY510 pKa = 10.29GGASRR515 pKa = 11.84QQQILLLRR523 pKa = 11.84SACDD527 pKa = 3.61IIVCTPGRR535 pKa = 3.69
MM1 pKa = 7.43SRR3 pKa = 11.84LGLSKK8 pKa = 10.9ALLRR12 pKa = 11.84FSLARR17 pKa = 11.84LRR19 pKa = 11.84PAATAAAAAATAATTAPAPSRR40 pKa = 11.84RR41 pKa = 11.84LLSSLSAAAPAAAAAGAAAAPAAAAGTEE69 pKa = 4.17TSPGVRR75 pKa = 11.84PPSLEE80 pKa = 3.74GKK82 pKa = 9.92FFTPAKK88 pKa = 9.09PWGGEE93 pKa = 3.77QQQQQQQQQHH103 pKa = 5.7LQQQQHH109 pKa = 5.59LHH111 pKa = 4.86QQQQHH116 pKa = 5.29LHH118 pKa = 4.74QQQQHH123 pKa = 5.27FHH125 pKa = 5.05QQQHH129 pKa = 5.48LHH131 pKa = 5.15QQQQLHH137 pKa = 5.12QQQQHH142 pKa = 5.13LHH144 pKa = 5.42HH145 pKa = 6.71HH146 pKa = 5.28QQQLHH151 pKa = 5.94HH152 pKa = 5.79QQQLHH157 pKa = 4.55QQQHH161 pKa = 5.23LHH163 pKa = 5.08QQQQLHH169 pKa = 6.12HH170 pKa = 5.63QQQLHH175 pKa = 4.81QQQQHH180 pKa = 5.34LHH182 pKa = 4.7QQQHH186 pKa = 5.41LHH188 pKa = 4.98QQQHH192 pKa = 5.81LQQQLQQQAQQQQQRR207 pKa = 11.84GRR209 pKa = 11.84GAFLPADD216 pKa = 4.31DD217 pKa = 4.56PWYY220 pKa = 9.76SSSYY224 pKa = 10.26EE225 pKa = 4.04PVSPPPRR232 pKa = 11.84QQQQQQQQQQQQQHH246 pKa = 5.16WRR248 pKa = 11.84PSAATAAAGGPGAGLRR264 pKa = 11.84PIDD267 pKa = 3.63WSKK270 pKa = 11.82KK271 pKa = 7.87NLPPIYY277 pKa = 9.33PVAPPPPPAAAAAAAAPGAAAAAAAAAATSAARR310 pKa = 11.84MKK312 pKa = 10.54EE313 pKa = 3.81LGIKK317 pKa = 9.91IEE319 pKa = 4.48GNAPLPLLLPASLPAAFAAAGLPDD343 pKa = 4.77LLLHH347 pKa = 7.04ALQHH351 pKa = 6.73LPQQQQQQQQQQQQQQQQISTLFPIQQIGWSCCFTGRR388 pKa = 11.84DD389 pKa = 4.19LIAIAQTGSGKK400 pKa = 9.22TIGYY404 pKa = 7.78LIPALIHH411 pKa = 6.94LLRR414 pKa = 11.84QQQQQQLQQQQQQQQGNSSSSSSSSSSRR442 pKa = 11.84GPTVLIVGPTRR453 pKa = 11.84EE454 pKa = 4.32LVQQIHH460 pKa = 6.5AEE462 pKa = 4.06CQRR465 pKa = 11.84ILKK468 pKa = 9.82AAAAAAAARR477 pKa = 11.84ASAAAAGGTATAAAAAAAAAAANTPLARR505 pKa = 11.84CVAVYY510 pKa = 10.29GGASRR515 pKa = 11.84QQQILLLRR523 pKa = 11.84SACDD527 pKa = 3.61IIVCTPGRR535 pKa = 3.69
Molecular weight: 57.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4025948 |
9 |
9976 |
664.7 |
71.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.095 ± 0.084 | 1.781 ± 0.02 |
3.887 ± 0.022 | 6.32 ± 0.04 |
2.384 ± 0.017 | 6.412 ± 0.042 |
2.17 ± 0.014 | 2.733 ± 0.019 |
3.647 ± 0.027 | 9.215 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.43 ± 0.012 | 2.65 ± 0.018 |
5.725 ± 0.035 | 10.399 ± 0.1 |
5.638 ± 0.028 | 9.449 ± 0.049 |
4.819 ± 0.021 | 4.808 ± 0.027 |
0.913 ± 0.008 | 1.525 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
