
human papillomavirus 200
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 2
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
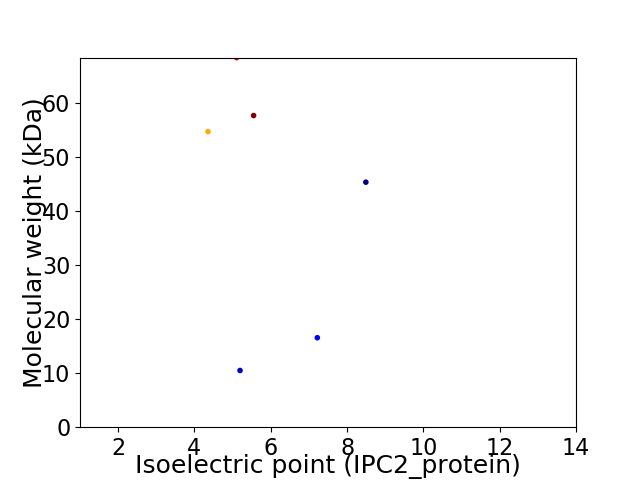
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H4M3P4|A0A0H4M3P4_9PAPI Minor capsid protein L2 OS=human papillomavirus 200 OX=1682339 GN=L2 PE=3 SV=1
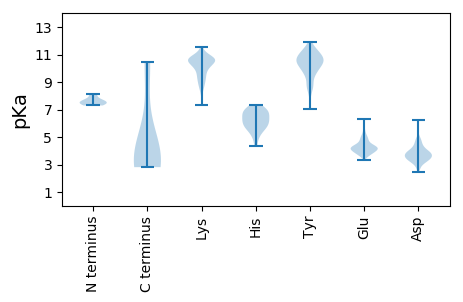
MM1 pKa = 7.43SLPRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.58RR8 pKa = 11.84ASPTDD13 pKa = 3.72LYY15 pKa = 10.02KK16 pKa = 10.6TCLQGGDD23 pKa = 4.92CIPDD27 pKa = 3.32VKK29 pKa = 11.21NKK31 pKa = 10.3FEE33 pKa = 4.15NTTIADD39 pKa = 3.72WLLKK43 pKa = 10.07IFGSIVYY50 pKa = 9.49FGNLGIGSGKK60 pKa = 8.45GTGGSFGYY68 pKa = 10.22RR69 pKa = 11.84PLGSAGSGRR78 pKa = 11.84PAQEE82 pKa = 4.23LPITRR87 pKa = 11.84PNVVIEE93 pKa = 4.36PIGPQSIVPIDD104 pKa = 4.02PGASSIVPLAEE115 pKa = 4.87GGLDD119 pKa = 3.36TSFITPDD126 pKa = 3.31AGPGVGGEE134 pKa = 4.29DD135 pKa = 2.9IEE137 pKa = 5.81LFTFTDD143 pKa = 3.7PATDD147 pKa = 3.16VGGVSGGPHH156 pKa = 6.92IISTQEE162 pKa = 3.58NEE164 pKa = 3.99TAIIDD169 pKa = 3.98ASPTVPPRR177 pKa = 11.84QVLYY181 pKa = 10.76DD182 pKa = 3.8SYY184 pKa = 11.73SNTLFEE190 pKa = 4.24THH192 pKa = 6.4VNPFLNNSLNTTNIFVDD209 pKa = 3.85PQLAGDD215 pKa = 3.96TVGEE219 pKa = 4.34NIFEE223 pKa = 4.72NIPLEE228 pKa = 4.33DD229 pKa = 4.79LSLQTLPTEE238 pKa = 4.32STPIRR243 pKa = 11.84PGRR246 pKa = 11.84ILRR249 pKa = 11.84TPAQRR254 pKa = 11.84SYY256 pKa = 11.21NRR258 pKa = 11.84FMEE261 pKa = 4.09QYY263 pKa = 9.76PIQSLDD269 pKa = 3.64FLSQPSRR276 pKa = 11.84LVQFEE281 pKa = 4.31FEE283 pKa = 4.7NPAFDD288 pKa = 4.68PDD290 pKa = 3.26ISLEE294 pKa = 3.87FQRR297 pKa = 11.84DD298 pKa = 3.77VNRR301 pKa = 11.84LEE303 pKa = 4.32AAPNAAFADD312 pKa = 3.47IAYY315 pKa = 9.96LSRR318 pKa = 11.84PRR320 pKa = 11.84VSLTSEE326 pKa = 3.67GLVRR330 pKa = 11.84VSRR333 pKa = 11.84IGTRR337 pKa = 11.84AALQTRR343 pKa = 11.84SGLTVGPQVHH353 pKa = 5.92YY354 pKa = 10.74FMDD357 pKa = 4.47LSAIPSEE364 pKa = 4.35NIEE367 pKa = 4.21LQPLGDD373 pKa = 3.65FTSRR377 pKa = 11.84STVVDD382 pKa = 4.01DD383 pKa = 3.95YY384 pKa = 11.94LAVTEE389 pKa = 5.01IDD391 pKa = 3.89DD392 pKa = 4.26PANIANVTYY401 pKa = 10.85SEE403 pKa = 5.44DD404 pKa = 3.9DD405 pKa = 3.78LLDD408 pKa = 4.29PLLEE412 pKa = 4.52SFDD415 pKa = 3.96NSHH418 pKa = 6.27ITIQGVNEE426 pKa = 4.19EE427 pKa = 4.64GDD429 pKa = 4.21TVTLPLPSVSNASKK443 pKa = 10.75IFIADD448 pKa = 4.33LANGLIFNDD457 pKa = 4.44YY458 pKa = 10.69NPQIYY463 pKa = 8.34PASTVISGSPWYY475 pKa = 10.6SIIEE479 pKa = 4.01RR480 pKa = 11.84NFADD484 pKa = 4.79FVLDD488 pKa = 3.51PAFIPRR494 pKa = 11.84KK495 pKa = 9.12RR496 pKa = 11.84RR497 pKa = 11.84RR498 pKa = 11.84LEE500 pKa = 3.57ILL502 pKa = 3.33
MM1 pKa = 7.43SLPRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.58RR8 pKa = 11.84ASPTDD13 pKa = 3.72LYY15 pKa = 10.02KK16 pKa = 10.6TCLQGGDD23 pKa = 4.92CIPDD27 pKa = 3.32VKK29 pKa = 11.21NKK31 pKa = 10.3FEE33 pKa = 4.15NTTIADD39 pKa = 3.72WLLKK43 pKa = 10.07IFGSIVYY50 pKa = 9.49FGNLGIGSGKK60 pKa = 8.45GTGGSFGYY68 pKa = 10.22RR69 pKa = 11.84PLGSAGSGRR78 pKa = 11.84PAQEE82 pKa = 4.23LPITRR87 pKa = 11.84PNVVIEE93 pKa = 4.36PIGPQSIVPIDD104 pKa = 4.02PGASSIVPLAEE115 pKa = 4.87GGLDD119 pKa = 3.36TSFITPDD126 pKa = 3.31AGPGVGGEE134 pKa = 4.29DD135 pKa = 2.9IEE137 pKa = 5.81LFTFTDD143 pKa = 3.7PATDD147 pKa = 3.16VGGVSGGPHH156 pKa = 6.92IISTQEE162 pKa = 3.58NEE164 pKa = 3.99TAIIDD169 pKa = 3.98ASPTVPPRR177 pKa = 11.84QVLYY181 pKa = 10.76DD182 pKa = 3.8SYY184 pKa = 11.73SNTLFEE190 pKa = 4.24THH192 pKa = 6.4VNPFLNNSLNTTNIFVDD209 pKa = 3.85PQLAGDD215 pKa = 3.96TVGEE219 pKa = 4.34NIFEE223 pKa = 4.72NIPLEE228 pKa = 4.33DD229 pKa = 4.79LSLQTLPTEE238 pKa = 4.32STPIRR243 pKa = 11.84PGRR246 pKa = 11.84ILRR249 pKa = 11.84TPAQRR254 pKa = 11.84SYY256 pKa = 11.21NRR258 pKa = 11.84FMEE261 pKa = 4.09QYY263 pKa = 9.76PIQSLDD269 pKa = 3.64FLSQPSRR276 pKa = 11.84LVQFEE281 pKa = 4.31FEE283 pKa = 4.7NPAFDD288 pKa = 4.68PDD290 pKa = 3.26ISLEE294 pKa = 3.87FQRR297 pKa = 11.84DD298 pKa = 3.77VNRR301 pKa = 11.84LEE303 pKa = 4.32AAPNAAFADD312 pKa = 3.47IAYY315 pKa = 9.96LSRR318 pKa = 11.84PRR320 pKa = 11.84VSLTSEE326 pKa = 3.67GLVRR330 pKa = 11.84VSRR333 pKa = 11.84IGTRR337 pKa = 11.84AALQTRR343 pKa = 11.84SGLTVGPQVHH353 pKa = 5.92YY354 pKa = 10.74FMDD357 pKa = 4.47LSAIPSEE364 pKa = 4.35NIEE367 pKa = 4.21LQPLGDD373 pKa = 3.65FTSRR377 pKa = 11.84STVVDD382 pKa = 4.01DD383 pKa = 3.95YY384 pKa = 11.94LAVTEE389 pKa = 5.01IDD391 pKa = 3.89DD392 pKa = 4.26PANIANVTYY401 pKa = 10.85SEE403 pKa = 5.44DD404 pKa = 3.9DD405 pKa = 3.78LLDD408 pKa = 4.29PLLEE412 pKa = 4.52SFDD415 pKa = 3.96NSHH418 pKa = 6.27ITIQGVNEE426 pKa = 4.19EE427 pKa = 4.64GDD429 pKa = 4.21TVTLPLPSVSNASKK443 pKa = 10.75IFIADD448 pKa = 4.33LANGLIFNDD457 pKa = 4.44YY458 pKa = 10.69NPQIYY463 pKa = 8.34PASTVISGSPWYY475 pKa = 10.6SIIEE479 pKa = 4.01RR480 pKa = 11.84NFADD484 pKa = 4.79FVLDD488 pKa = 3.51PAFIPRR494 pKa = 11.84KK495 pKa = 9.12RR496 pKa = 11.84RR497 pKa = 11.84RR498 pKa = 11.84LEE500 pKa = 3.57ILL502 pKa = 3.33
Molecular weight: 54.67 kDa
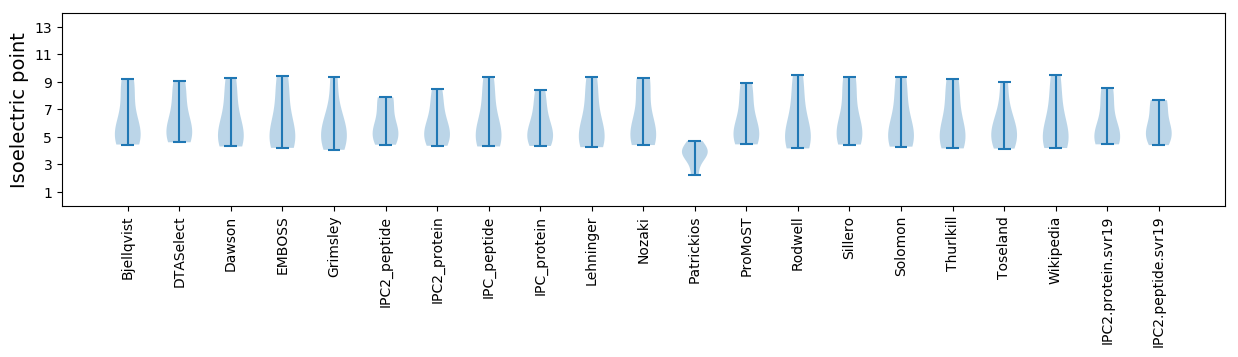
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H4LNX5|A0A0H4LNX5_9PAPI Protein E7 OS=human papillomavirus 200 OX=1682339 GN=E7 PE=3 SV=1
MM1 pKa = 7.3QPEE4 pKa = 4.41TQEE7 pKa = 4.06SLSARR12 pKa = 11.84FAAQQEE18 pKa = 4.31IQLNLIEE25 pKa = 4.82KK26 pKa = 10.06EE27 pKa = 4.2SYY29 pKa = 10.16DD30 pKa = 4.07LKK32 pKa = 11.44DD33 pKa = 3.25HH34 pKa = 6.74LEE36 pKa = 3.72YY37 pKa = 10.32WKK39 pKa = 10.64AVRR42 pKa = 11.84AEE44 pKa = 3.99NVIAYY49 pKa = 7.06YY50 pKa = 10.49ARR52 pKa = 11.84KK53 pKa = 8.79EE54 pKa = 4.21HH55 pKa = 6.44INKK58 pKa = 10.1LGLQQLPTTAVTEE71 pKa = 4.42YY72 pKa = 10.29KK73 pKa = 10.6AKK75 pKa = 10.43EE76 pKa = 4.31AINMQLLIQSLMKK89 pKa = 10.58SSFAMEE95 pKa = 3.99RR96 pKa = 11.84WTLSEE101 pKa = 4.11TSAEE105 pKa = 4.64SINSSPRR112 pKa = 11.84NCFKK116 pKa = 10.81KK117 pKa = 10.53SPFIVTVWFDD127 pKa = 3.16NDD129 pKa = 3.18EE130 pKa = 4.52RR131 pKa = 11.84NSFPYY136 pKa = 9.67TCWDD140 pKa = 3.45YY141 pKa = 11.35IYY143 pKa = 10.76FQDD146 pKa = 5.01EE147 pKa = 3.69QNMWHH152 pKa = 6.32KK153 pKa = 8.96THH155 pKa = 6.74GLVDD159 pKa = 3.67HH160 pKa = 7.32DD161 pKa = 3.6GCYY164 pKa = 10.81YY165 pKa = 10.71KK166 pKa = 10.87DD167 pKa = 3.67NNGDD171 pKa = 3.29SVYY174 pKa = 10.66FILFHH179 pKa = 7.04PDD181 pKa = 2.47AVKK184 pKa = 10.9YY185 pKa = 10.89GNTGLWTVKK194 pKa = 10.17FKK196 pKa = 11.52NKK198 pKa = 9.19TISASVTSSSRR209 pKa = 11.84HH210 pKa = 5.59SIPSFEE216 pKa = 3.86GRR218 pKa = 11.84VWPSTSTSPEE228 pKa = 4.0SPGGGLQDD236 pKa = 3.62TSQNSSTEE244 pKa = 4.31SPTSSTPGLRR254 pKa = 11.84LRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84RR260 pKa = 11.84QSGEE264 pKa = 3.65PGEE267 pKa = 4.97SGTADD272 pKa = 3.45EE273 pKa = 4.58TPKK276 pKa = 10.6RR277 pKa = 11.84RR278 pKa = 11.84GIKK281 pKa = 9.89RR282 pKa = 11.84KK283 pKa = 9.73LGSDD287 pKa = 3.56TAPSPSEE294 pKa = 3.88VGSRR298 pKa = 11.84SHH300 pKa = 4.87TLEE303 pKa = 3.63RR304 pKa = 11.84SGYY307 pKa = 8.52SRR309 pKa = 11.84LRR311 pKa = 11.84RR312 pKa = 11.84LQEE315 pKa = 3.72EE316 pKa = 4.18ARR318 pKa = 11.84DD319 pKa = 3.7PPIALFTGHH328 pKa = 6.24QNNLKK333 pKa = 9.88CWRR336 pKa = 11.84NRR338 pKa = 11.84CTTKK342 pKa = 10.65YY343 pKa = 10.59SSLFVAFSSVWKK355 pKa = 9.45WLGPNVSGGAAKK367 pKa = 10.64LLVAFSSNSQRR378 pKa = 11.84EE379 pKa = 4.31LFLSTVHH386 pKa = 6.07IPKK389 pKa = 8.88GTSVTLGNLNSLL401 pKa = 4.15
MM1 pKa = 7.3QPEE4 pKa = 4.41TQEE7 pKa = 4.06SLSARR12 pKa = 11.84FAAQQEE18 pKa = 4.31IQLNLIEE25 pKa = 4.82KK26 pKa = 10.06EE27 pKa = 4.2SYY29 pKa = 10.16DD30 pKa = 4.07LKK32 pKa = 11.44DD33 pKa = 3.25HH34 pKa = 6.74LEE36 pKa = 3.72YY37 pKa = 10.32WKK39 pKa = 10.64AVRR42 pKa = 11.84AEE44 pKa = 3.99NVIAYY49 pKa = 7.06YY50 pKa = 10.49ARR52 pKa = 11.84KK53 pKa = 8.79EE54 pKa = 4.21HH55 pKa = 6.44INKK58 pKa = 10.1LGLQQLPTTAVTEE71 pKa = 4.42YY72 pKa = 10.29KK73 pKa = 10.6AKK75 pKa = 10.43EE76 pKa = 4.31AINMQLLIQSLMKK89 pKa = 10.58SSFAMEE95 pKa = 3.99RR96 pKa = 11.84WTLSEE101 pKa = 4.11TSAEE105 pKa = 4.64SINSSPRR112 pKa = 11.84NCFKK116 pKa = 10.81KK117 pKa = 10.53SPFIVTVWFDD127 pKa = 3.16NDD129 pKa = 3.18EE130 pKa = 4.52RR131 pKa = 11.84NSFPYY136 pKa = 9.67TCWDD140 pKa = 3.45YY141 pKa = 11.35IYY143 pKa = 10.76FQDD146 pKa = 5.01EE147 pKa = 3.69QNMWHH152 pKa = 6.32KK153 pKa = 8.96THH155 pKa = 6.74GLVDD159 pKa = 3.67HH160 pKa = 7.32DD161 pKa = 3.6GCYY164 pKa = 10.81YY165 pKa = 10.71KK166 pKa = 10.87DD167 pKa = 3.67NNGDD171 pKa = 3.29SVYY174 pKa = 10.66FILFHH179 pKa = 7.04PDD181 pKa = 2.47AVKK184 pKa = 10.9YY185 pKa = 10.89GNTGLWTVKK194 pKa = 10.17FKK196 pKa = 11.52NKK198 pKa = 9.19TISASVTSSSRR209 pKa = 11.84HH210 pKa = 5.59SIPSFEE216 pKa = 3.86GRR218 pKa = 11.84VWPSTSTSPEE228 pKa = 4.0SPGGGLQDD236 pKa = 3.62TSQNSSTEE244 pKa = 4.31SPTSSTPGLRR254 pKa = 11.84LRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84RR260 pKa = 11.84QSGEE264 pKa = 3.65PGEE267 pKa = 4.97SGTADD272 pKa = 3.45EE273 pKa = 4.58TPKK276 pKa = 10.6RR277 pKa = 11.84RR278 pKa = 11.84GIKK281 pKa = 9.89RR282 pKa = 11.84KK283 pKa = 9.73LGSDD287 pKa = 3.56TAPSPSEE294 pKa = 3.88VGSRR298 pKa = 11.84SHH300 pKa = 4.87TLEE303 pKa = 3.63RR304 pKa = 11.84SGYY307 pKa = 8.52SRR309 pKa = 11.84LRR311 pKa = 11.84RR312 pKa = 11.84LQEE315 pKa = 3.72EE316 pKa = 4.18ARR318 pKa = 11.84DD319 pKa = 3.7PPIALFTGHH328 pKa = 6.24QNNLKK333 pKa = 9.88CWRR336 pKa = 11.84NRR338 pKa = 11.84CTTKK342 pKa = 10.65YY343 pKa = 10.59SSLFVAFSSVWKK355 pKa = 9.45WLGPNVSGGAAKK367 pKa = 10.64LLVAFSSNSQRR378 pKa = 11.84EE379 pKa = 4.31LFLSTVHH386 pKa = 6.07IPKK389 pKa = 8.88GTSVTLGNLNSLL401 pKa = 4.15
Molecular weight: 45.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2247 |
93 |
598 |
374.5 |
42.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.919 ± 0.277 | 2.403 ± 0.849 |
6.008 ± 0.474 | 6.142 ± 0.312 |
5.34 ± 0.452 | 5.518 ± 0.884 |
1.38 ± 0.302 | 5.518 ± 0.655 |
5.296 ± 0.97 | 9.39 ± 0.466 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.424 ± 0.271 | 5.696 ± 0.235 |
5.652 ± 0.903 | 3.961 ± 0.199 |
5.118 ± 0.484 | 8.767 ± 0.848 |
6.186 ± 0.35 | 5.385 ± 0.302 |
1.424 ± 0.284 | 3.471 ± 0.298 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
