
Aquamicrobium aerolatum DSM 21857
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Aquamicrobium; Aquamicrobium aerolatum
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
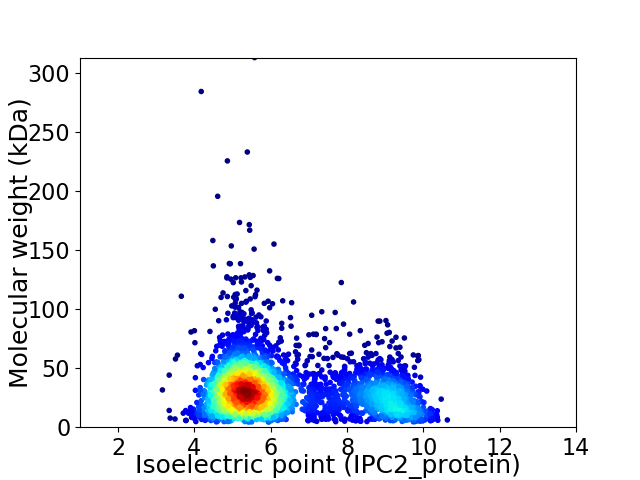
Virtual 2D-PAGE plot for 3495 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I3T2P5|A0A1I3T2P5_9RHIZ Sterol carrier protein OS=Aquamicrobium aerolatum DSM 21857 OX=1121003 GN=SAMN03080618_03537 PE=4 SV=1
MM1 pKa = 7.73ALTHH5 pKa = 6.75LSFDD9 pKa = 2.85ATYY12 pKa = 11.36YY13 pKa = 7.12MTEE16 pKa = 4.17RR17 pKa = 11.84PDD19 pKa = 3.35VLTAYY24 pKa = 10.19VNAGAEE30 pKa = 4.44TGTGMNWAQFAEE42 pKa = 3.84QHH44 pKa = 6.1YY45 pKa = 11.26NDD47 pKa = 4.76FGWKK51 pKa = 9.44EE52 pKa = 4.17GYY54 pKa = 9.71NPNAIFDD61 pKa = 3.85TSEE64 pKa = 3.73YY65 pKa = 10.24LAANIDD71 pKa = 3.8VLNAGVNPFQHH82 pKa = 6.19YY83 pKa = 9.81LQFGAYY89 pKa = 7.3EE90 pKa = 3.98KK91 pKa = 10.38RR92 pKa = 11.84APSDD96 pKa = 3.41SFISFEE102 pKa = 4.12DD103 pKa = 4.24FDD105 pKa = 3.87WEE107 pKa = 4.66TYY109 pKa = 10.55LGANSDD115 pKa = 3.73LTDD118 pKa = 3.72AGIEE122 pKa = 4.18TAEE125 pKa = 4.36DD126 pKa = 4.08AYY128 pKa = 10.86GHH130 pKa = 5.91YY131 pKa = 10.95VLFGQFEE138 pKa = 4.36VRR140 pKa = 11.84DD141 pKa = 4.23GKK143 pKa = 10.08PEE145 pKa = 3.74EE146 pKa = 5.2AIPSVPGEE154 pKa = 4.48TYY156 pKa = 10.57TLTTGVDD163 pKa = 3.05AGADD167 pKa = 3.65FTGTADD173 pKa = 3.38NDD175 pKa = 3.74TFRR178 pKa = 11.84AFDD181 pKa = 3.47MDD183 pKa = 4.39GPSGTAGATLQSWDD197 pKa = 4.15ILDD200 pKa = 4.14GGAGVDD206 pKa = 3.7TLNIATGAAADD217 pKa = 3.88NAAPTLRR224 pKa = 11.84NIEE227 pKa = 4.47IINNAHH233 pKa = 7.26LGQTINLASATGVQQIWTDD252 pKa = 3.25MTGFTGTAATRR263 pKa = 11.84YY264 pKa = 9.49NDD266 pKa = 3.4ASVATIFGIKK276 pKa = 9.4GAEE279 pKa = 4.12GSNSDD284 pKa = 4.09VNITFADD291 pKa = 3.77SLEE294 pKa = 4.25GDD296 pKa = 3.75TTVNFALEE304 pKa = 4.02GNAAGSYY311 pKa = 10.42AGFYY315 pKa = 10.98LEE317 pKa = 6.11DD318 pKa = 3.69EE319 pKa = 4.5GVEE322 pKa = 4.03NAVITVAEE330 pKa = 4.47GNGGFTTVSGVSSVTASGAGDD351 pKa = 3.43FGFYY355 pKa = 9.43TWDD358 pKa = 3.22NTTIEE363 pKa = 4.61SFNASAVTGDD373 pKa = 3.47VYY375 pKa = 10.98LTGAAGGTAAAFDD388 pKa = 4.07EE389 pKa = 4.83DD390 pKa = 4.84ANISSGAGNDD400 pKa = 3.47RR401 pKa = 11.84IYY403 pKa = 11.28VGNSDD408 pKa = 3.81GALTISTGAGNDD420 pKa = 3.63IIVSGSGNDD429 pKa = 3.77TIIAGAGKK437 pKa = 10.07NDD439 pKa = 3.52LTGGLGEE446 pKa = 4.53DD447 pKa = 4.01TFVLDD452 pKa = 4.48IKK454 pKa = 10.67GTVLGSLDD462 pKa = 4.05VINDD466 pKa = 3.42FNFGDD471 pKa = 3.96AQDD474 pKa = 3.68TLVFGEE480 pKa = 4.65TEE482 pKa = 3.91LTNDD486 pKa = 2.99NFASVGIAVSYY497 pKa = 11.26NDD499 pKa = 4.68LRR501 pKa = 11.84EE502 pKa = 4.05LAQGAFSDD510 pKa = 4.36DD511 pKa = 3.41VSFVAGTFGADD522 pKa = 2.51TYY524 pKa = 11.4VFHH527 pKa = 7.76DD528 pKa = 3.94ADD530 pKa = 4.14ADD532 pKa = 4.13GVADD536 pKa = 4.41AAVKK540 pKa = 10.7LIGTGSLLGADD551 pKa = 4.1AFEE554 pKa = 4.47VAAVV558 pKa = 3.41
MM1 pKa = 7.73ALTHH5 pKa = 6.75LSFDD9 pKa = 2.85ATYY12 pKa = 11.36YY13 pKa = 7.12MTEE16 pKa = 4.17RR17 pKa = 11.84PDD19 pKa = 3.35VLTAYY24 pKa = 10.19VNAGAEE30 pKa = 4.44TGTGMNWAQFAEE42 pKa = 3.84QHH44 pKa = 6.1YY45 pKa = 11.26NDD47 pKa = 4.76FGWKK51 pKa = 9.44EE52 pKa = 4.17GYY54 pKa = 9.71NPNAIFDD61 pKa = 3.85TSEE64 pKa = 3.73YY65 pKa = 10.24LAANIDD71 pKa = 3.8VLNAGVNPFQHH82 pKa = 6.19YY83 pKa = 9.81LQFGAYY89 pKa = 7.3EE90 pKa = 3.98KK91 pKa = 10.38RR92 pKa = 11.84APSDD96 pKa = 3.41SFISFEE102 pKa = 4.12DD103 pKa = 4.24FDD105 pKa = 3.87WEE107 pKa = 4.66TYY109 pKa = 10.55LGANSDD115 pKa = 3.73LTDD118 pKa = 3.72AGIEE122 pKa = 4.18TAEE125 pKa = 4.36DD126 pKa = 4.08AYY128 pKa = 10.86GHH130 pKa = 5.91YY131 pKa = 10.95VLFGQFEE138 pKa = 4.36VRR140 pKa = 11.84DD141 pKa = 4.23GKK143 pKa = 10.08PEE145 pKa = 3.74EE146 pKa = 5.2AIPSVPGEE154 pKa = 4.48TYY156 pKa = 10.57TLTTGVDD163 pKa = 3.05AGADD167 pKa = 3.65FTGTADD173 pKa = 3.38NDD175 pKa = 3.74TFRR178 pKa = 11.84AFDD181 pKa = 3.47MDD183 pKa = 4.39GPSGTAGATLQSWDD197 pKa = 4.15ILDD200 pKa = 4.14GGAGVDD206 pKa = 3.7TLNIATGAAADD217 pKa = 3.88NAAPTLRR224 pKa = 11.84NIEE227 pKa = 4.47IINNAHH233 pKa = 7.26LGQTINLASATGVQQIWTDD252 pKa = 3.25MTGFTGTAATRR263 pKa = 11.84YY264 pKa = 9.49NDD266 pKa = 3.4ASVATIFGIKK276 pKa = 9.4GAEE279 pKa = 4.12GSNSDD284 pKa = 4.09VNITFADD291 pKa = 3.77SLEE294 pKa = 4.25GDD296 pKa = 3.75TTVNFALEE304 pKa = 4.02GNAAGSYY311 pKa = 10.42AGFYY315 pKa = 10.98LEE317 pKa = 6.11DD318 pKa = 3.69EE319 pKa = 4.5GVEE322 pKa = 4.03NAVITVAEE330 pKa = 4.47GNGGFTTVSGVSSVTASGAGDD351 pKa = 3.43FGFYY355 pKa = 9.43TWDD358 pKa = 3.22NTTIEE363 pKa = 4.61SFNASAVTGDD373 pKa = 3.47VYY375 pKa = 10.98LTGAAGGTAAAFDD388 pKa = 4.07EE389 pKa = 4.83DD390 pKa = 4.84ANISSGAGNDD400 pKa = 3.47RR401 pKa = 11.84IYY403 pKa = 11.28VGNSDD408 pKa = 3.81GALTISTGAGNDD420 pKa = 3.63IIVSGSGNDD429 pKa = 3.77TIIAGAGKK437 pKa = 10.07NDD439 pKa = 3.52LTGGLGEE446 pKa = 4.53DD447 pKa = 4.01TFVLDD452 pKa = 4.48IKK454 pKa = 10.67GTVLGSLDD462 pKa = 4.05VINDD466 pKa = 3.42FNFGDD471 pKa = 3.96AQDD474 pKa = 3.68TLVFGEE480 pKa = 4.65TEE482 pKa = 3.91LTNDD486 pKa = 2.99NFASVGIAVSYY497 pKa = 11.26NDD499 pKa = 4.68LRR501 pKa = 11.84EE502 pKa = 4.05LAQGAFSDD510 pKa = 4.36DD511 pKa = 3.41VSFVAGTFGADD522 pKa = 2.51TYY524 pKa = 11.4VFHH527 pKa = 7.76DD528 pKa = 3.94ADD530 pKa = 4.14ADD532 pKa = 4.13GVADD536 pKa = 4.41AAVKK540 pKa = 10.7LIGTGSLLGADD551 pKa = 4.1AFEE554 pKa = 4.47VAAVV558 pKa = 3.41
Molecular weight: 57.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I3J2R1|A0A1I3J2R1_9RHIZ Purine-binding chemotaxis protein CheW OS=Aquamicrobium aerolatum DSM 21857 OX=1121003 GN=SAMN03080618_00763 PE=4 SV=1
MM1 pKa = 7.31FCSTMRR7 pKa = 11.84SISNNHH13 pKa = 4.8VRR15 pKa = 11.84SEE17 pKa = 3.69ADD19 pKa = 3.08IIVVRR24 pKa = 11.84QRR26 pKa = 11.84PGHH29 pKa = 6.36PSQGLLIWGNTVFPCALGRR48 pKa = 11.84GGIKK52 pKa = 10.07ALKK55 pKa = 10.16RR56 pKa = 11.84EE57 pKa = 4.24GDD59 pKa = 3.46GATPLGSTRR68 pKa = 11.84LLYY71 pKa = 10.54GWFRR75 pKa = 11.84HH76 pKa = 5.29GKK78 pKa = 8.55IEE80 pKa = 4.32SRR82 pKa = 11.84RR83 pKa = 11.84SGLALRR89 pKa = 11.84QITPSDD95 pKa = 3.74GWCDD99 pKa = 3.36APADD103 pKa = 4.2RR104 pKa = 11.84NYY106 pKa = 10.59NRR108 pKa = 11.84PVTLPYY114 pKa = 9.5PASSEE119 pKa = 3.97TMEE122 pKa = 4.6RR123 pKa = 11.84KK124 pKa = 9.79DD125 pKa = 4.65RR126 pKa = 11.84LYY128 pKa = 10.92DD129 pKa = 3.26CCIVMDD135 pKa = 3.86YY136 pKa = 10.78NIRR139 pKa = 11.84PRR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84GRR145 pKa = 11.84GSAIFFHH152 pKa = 6.63IAKK155 pKa = 9.69PGFPPTQGCIAVEE168 pKa = 3.93PQVMQRR174 pKa = 11.84LLPHH178 pKa = 6.75LSPRR182 pKa = 11.84TRR184 pKa = 11.84IKK186 pKa = 10.59VVRR189 pKa = 3.95
MM1 pKa = 7.31FCSTMRR7 pKa = 11.84SISNNHH13 pKa = 4.8VRR15 pKa = 11.84SEE17 pKa = 3.69ADD19 pKa = 3.08IIVVRR24 pKa = 11.84QRR26 pKa = 11.84PGHH29 pKa = 6.36PSQGLLIWGNTVFPCALGRR48 pKa = 11.84GGIKK52 pKa = 10.07ALKK55 pKa = 10.16RR56 pKa = 11.84EE57 pKa = 4.24GDD59 pKa = 3.46GATPLGSTRR68 pKa = 11.84LLYY71 pKa = 10.54GWFRR75 pKa = 11.84HH76 pKa = 5.29GKK78 pKa = 8.55IEE80 pKa = 4.32SRR82 pKa = 11.84RR83 pKa = 11.84SGLALRR89 pKa = 11.84QITPSDD95 pKa = 3.74GWCDD99 pKa = 3.36APADD103 pKa = 4.2RR104 pKa = 11.84NYY106 pKa = 10.59NRR108 pKa = 11.84PVTLPYY114 pKa = 9.5PASSEE119 pKa = 3.97TMEE122 pKa = 4.6RR123 pKa = 11.84KK124 pKa = 9.79DD125 pKa = 4.65RR126 pKa = 11.84LYY128 pKa = 10.92DD129 pKa = 3.26CCIVMDD135 pKa = 3.86YY136 pKa = 10.78NIRR139 pKa = 11.84PRR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84GRR145 pKa = 11.84GSAIFFHH152 pKa = 6.63IAKK155 pKa = 9.69PGFPPTQGCIAVEE168 pKa = 3.93PQVMQRR174 pKa = 11.84LLPHH178 pKa = 6.75LSPRR182 pKa = 11.84TRR184 pKa = 11.84IKK186 pKa = 10.59VVRR189 pKa = 3.95
Molecular weight: 21.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1083778 |
39 |
2826 |
310.1 |
33.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.05 ± 0.055 | 0.772 ± 0.011 |
5.758 ± 0.032 | 5.933 ± 0.041 |
3.855 ± 0.029 | 8.311 ± 0.04 |
2.054 ± 0.022 | 5.579 ± 0.034 |
3.325 ± 0.032 | 9.854 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.704 ± 0.022 | 2.795 ± 0.023 |
4.824 ± 0.027 | 3.244 ± 0.025 |
6.909 ± 0.041 | 5.632 ± 0.025 |
5.319 ± 0.031 | 7.531 ± 0.035 |
1.275 ± 0.018 | 2.275 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
