
Aquimarina sp. TRL1
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Aquimarina; unclassified Aquimarina
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
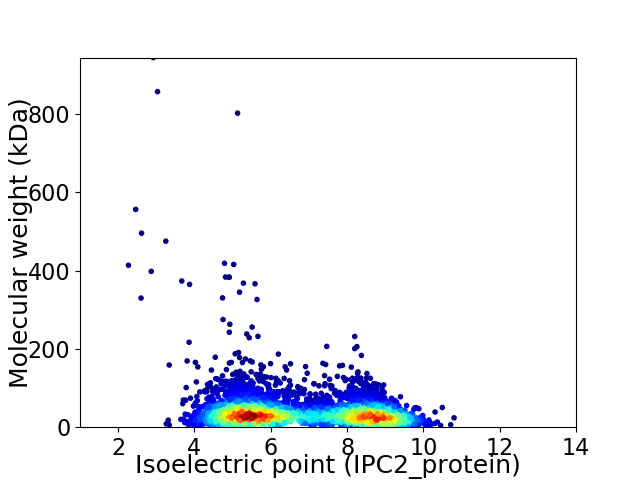
Virtual 2D-PAGE plot for 4415 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7H8PPI0|A0A7H8PPI0_9FLAO Ribonuclease Z OS=Aquimarina sp. TRL1 OX=2736252 GN=rnz PE=3 SV=1
MM1 pKa = 7.73KK2 pKa = 9.14KK3 pKa = 6.75TTYY6 pKa = 10.96LLIVFLGLLYY16 pKa = 10.23PKK18 pKa = 10.06FVSAQCEE25 pKa = 4.17VEE27 pKa = 4.35EE28 pKa = 4.4TFSVCDD34 pKa = 3.82MEE36 pKa = 5.6TVDD39 pKa = 5.0HH40 pKa = 7.01DD41 pKa = 4.54TNGTPDD47 pKa = 4.37GIINLFDD54 pKa = 3.39EE55 pKa = 5.05FQNVTGQSLTGGTWSIVPKK74 pKa = 10.32YY75 pKa = 10.57ASNLNPVTGEE85 pKa = 4.01VNVWAFPNSTVTDD98 pKa = 3.66TLEE101 pKa = 4.07EE102 pKa = 4.39YY103 pKa = 10.94VFEE106 pKa = 5.21FNTPACGDD114 pKa = 3.59TPAVIFTMILGPFSGVALPLGANNANIEE142 pKa = 3.99ICDD145 pKa = 3.88TDD147 pKa = 3.85TLNLFEE153 pKa = 5.69ALTSNEE159 pKa = 4.79SIPAPHH165 pKa = 7.24LNGTWEE171 pKa = 4.72FISNSSGAAFTGTFTLNDD189 pKa = 3.55HH190 pKa = 6.48SFSADD195 pKa = 2.79IPYY198 pKa = 10.28EE199 pKa = 3.81ITIPPINRR207 pKa = 11.84EE208 pKa = 3.89IFEE211 pKa = 4.14LRR213 pKa = 11.84YY214 pKa = 9.42VVPGMTPCATSEE226 pKa = 3.99EE227 pKa = 4.4TRR229 pKa = 11.84VRR231 pKa = 11.84VSVVRR236 pKa = 11.84QVSAGIMEE244 pKa = 5.24AGLICEE250 pKa = 4.27GDD252 pKa = 3.65ILAGLHH258 pKa = 6.77DD259 pKa = 5.07ADD261 pKa = 4.39IDD263 pKa = 4.21LRR265 pKa = 11.84DD266 pKa = 3.77DD267 pKa = 3.38QFLRR271 pKa = 11.84DD272 pKa = 3.81EE273 pKa = 4.78DD274 pKa = 4.51VGGTWSSQGADD285 pKa = 3.48PTNQISDD292 pKa = 3.84EE293 pKa = 3.98QDD295 pKa = 3.02SVINLRR301 pKa = 11.84EE302 pKa = 3.86IYY304 pKa = 10.76DD305 pKa = 3.64NVINGGNNLRR315 pKa = 11.84FGCVDD320 pKa = 3.98FGFTYY325 pKa = 10.26SVKK328 pKa = 10.16KK329 pKa = 10.44RR330 pKa = 11.84SAVCEE335 pKa = 4.3DD336 pKa = 3.87DD337 pKa = 5.23DD338 pKa = 3.93EE339 pKa = 5.05TVFFTFFEE347 pKa = 4.01QLRR350 pKa = 11.84PFNQAGEE357 pKa = 4.17IPEE360 pKa = 4.04VCANEE365 pKa = 4.2EE366 pKa = 4.26PGSIDD371 pKa = 5.36LFDD374 pKa = 4.76LLEE377 pKa = 4.08FTNEE381 pKa = 3.53NGIDD385 pKa = 4.22FIYY388 pKa = 11.1NDD390 pKa = 3.71DD391 pKa = 4.89AYY393 pKa = 11.26VKK395 pKa = 9.45WRR397 pKa = 11.84MVSGPSDD404 pKa = 3.58LPLSEE409 pKa = 4.32DD410 pKa = 3.32HH411 pKa = 6.92KK412 pKa = 11.2GTVQTLGAASGTYY425 pKa = 9.34VFEE428 pKa = 4.83FGVSPKK434 pKa = 9.94INCPGGEE441 pKa = 4.1AFCDD445 pKa = 3.57PFAAEE450 pKa = 4.56GEE452 pKa = 4.28EE453 pKa = 4.5GHH455 pKa = 7.0CEE457 pKa = 4.11FPCGVLTTQVTIRR470 pKa = 11.84VLEE473 pKa = 3.85FDD475 pKa = 4.07YY476 pKa = 11.32AGEE479 pKa = 4.25DD480 pKa = 3.26TDD482 pKa = 5.92GINICISEE490 pKa = 4.08NQIDD494 pKa = 4.43LRR496 pKa = 11.84SLLKK500 pKa = 10.35IEE502 pKa = 4.59GGNTIVDD509 pKa = 3.48TGVWTNEE516 pKa = 3.57AGEE519 pKa = 4.61EE520 pKa = 3.87IDD522 pKa = 3.49NTFVFPEE529 pKa = 4.04ISSPEE534 pKa = 3.91AFRR537 pKa = 11.84FTYY540 pKa = 9.55TATSDD545 pKa = 3.47MGCIDD550 pKa = 3.72AATLEE555 pKa = 4.54FTIHH559 pKa = 6.74PLPVAGEE566 pKa = 4.42DD567 pKa = 3.7GTASVCSDD575 pKa = 3.42NLTVTLFDD583 pKa = 4.19HH584 pKa = 7.45LEE586 pKa = 4.37GNPDD590 pKa = 3.25TTGIWTGPNGYY601 pKa = 8.05TSTDD605 pKa = 2.81HH606 pKa = 7.54LGRR609 pKa = 11.84FDD611 pKa = 4.82QNDD614 pKa = 3.52DD615 pKa = 3.62TLPILGAGRR624 pKa = 11.84FRR626 pKa = 11.84YY627 pKa = 7.99EE628 pKa = 3.76VPGNAGCLDD637 pKa = 3.71SDD639 pKa = 3.83ASFVDD644 pKa = 4.35VIIIEE649 pKa = 4.28PEE651 pKa = 3.87NIGNDD656 pKa = 3.44RR657 pKa = 11.84AEE659 pKa = 4.43TFCKK663 pKa = 9.62IDD665 pKa = 3.12GRR667 pKa = 11.84VNLFSLLDD675 pKa = 3.81RR676 pKa = 11.84NTVRR680 pKa = 11.84TGTFEE685 pKa = 4.32DD686 pKa = 4.08TDD688 pKa = 3.63DD689 pKa = 3.75TGALNSEE696 pKa = 4.19GVVEE700 pKa = 6.23FEE702 pKa = 4.15TLTNGIYY709 pKa = 9.99NFRR712 pKa = 11.84YY713 pKa = 9.65IISNTAPCEE722 pKa = 3.96EE723 pKa = 4.46SSLNVAIQIVDD734 pKa = 3.79LPEE737 pKa = 4.26PQVGDD742 pKa = 3.52QEE744 pKa = 4.92FCILDD749 pKa = 3.51AARR752 pKa = 11.84LEE754 pKa = 4.87DD755 pKa = 3.79IVVDD759 pKa = 3.33VLNYY763 pKa = 9.71NWYY766 pKa = 6.93EE767 pKa = 4.08TLEE770 pKa = 4.09GTIPIITNPILLDD783 pKa = 3.49EE784 pKa = 4.25QVFYY788 pKa = 10.74IANVDD793 pKa = 3.63ANNCEE798 pKa = 4.17SEE800 pKa = 4.2RR801 pKa = 11.84VEE803 pKa = 4.16VRR805 pKa = 11.84INILNIGEE813 pKa = 4.29RR814 pKa = 11.84SAQGEE819 pKa = 4.73LCTLDD824 pKa = 3.85FQDD827 pKa = 5.29GVSPNNDD834 pKa = 3.47VINDD838 pKa = 3.82TFDD841 pKa = 4.14LVIEE845 pKa = 4.26DD846 pKa = 4.4VYY848 pKa = 11.16NIPEE852 pKa = 4.5AFPDD856 pKa = 3.64FDD858 pKa = 4.72LKK860 pKa = 10.86IFNRR864 pKa = 11.84YY865 pKa = 8.99GSMVYY870 pKa = 10.02EE871 pKa = 4.52GNADD875 pKa = 3.55TEE877 pKa = 4.38EE878 pKa = 4.1FRR880 pKa = 11.84GEE882 pKa = 4.21SNISLRR888 pKa = 11.84LGDD891 pKa = 4.92DD892 pKa = 4.15LPSGTYY898 pKa = 9.9FYY900 pKa = 10.74IFTPNFEE907 pKa = 5.05NNFPIQGSFYY917 pKa = 10.87LSRR920 pKa = 5.17
MM1 pKa = 7.73KK2 pKa = 9.14KK3 pKa = 6.75TTYY6 pKa = 10.96LLIVFLGLLYY16 pKa = 10.23PKK18 pKa = 10.06FVSAQCEE25 pKa = 4.17VEE27 pKa = 4.35EE28 pKa = 4.4TFSVCDD34 pKa = 3.82MEE36 pKa = 5.6TVDD39 pKa = 5.0HH40 pKa = 7.01DD41 pKa = 4.54TNGTPDD47 pKa = 4.37GIINLFDD54 pKa = 3.39EE55 pKa = 5.05FQNVTGQSLTGGTWSIVPKK74 pKa = 10.32YY75 pKa = 10.57ASNLNPVTGEE85 pKa = 4.01VNVWAFPNSTVTDD98 pKa = 3.66TLEE101 pKa = 4.07EE102 pKa = 4.39YY103 pKa = 10.94VFEE106 pKa = 5.21FNTPACGDD114 pKa = 3.59TPAVIFTMILGPFSGVALPLGANNANIEE142 pKa = 3.99ICDD145 pKa = 3.88TDD147 pKa = 3.85TLNLFEE153 pKa = 5.69ALTSNEE159 pKa = 4.79SIPAPHH165 pKa = 7.24LNGTWEE171 pKa = 4.72FISNSSGAAFTGTFTLNDD189 pKa = 3.55HH190 pKa = 6.48SFSADD195 pKa = 2.79IPYY198 pKa = 10.28EE199 pKa = 3.81ITIPPINRR207 pKa = 11.84EE208 pKa = 3.89IFEE211 pKa = 4.14LRR213 pKa = 11.84YY214 pKa = 9.42VVPGMTPCATSEE226 pKa = 3.99EE227 pKa = 4.4TRR229 pKa = 11.84VRR231 pKa = 11.84VSVVRR236 pKa = 11.84QVSAGIMEE244 pKa = 5.24AGLICEE250 pKa = 4.27GDD252 pKa = 3.65ILAGLHH258 pKa = 6.77DD259 pKa = 5.07ADD261 pKa = 4.39IDD263 pKa = 4.21LRR265 pKa = 11.84DD266 pKa = 3.77DD267 pKa = 3.38QFLRR271 pKa = 11.84DD272 pKa = 3.81EE273 pKa = 4.78DD274 pKa = 4.51VGGTWSSQGADD285 pKa = 3.48PTNQISDD292 pKa = 3.84EE293 pKa = 3.98QDD295 pKa = 3.02SVINLRR301 pKa = 11.84EE302 pKa = 3.86IYY304 pKa = 10.76DD305 pKa = 3.64NVINGGNNLRR315 pKa = 11.84FGCVDD320 pKa = 3.98FGFTYY325 pKa = 10.26SVKK328 pKa = 10.16KK329 pKa = 10.44RR330 pKa = 11.84SAVCEE335 pKa = 4.3DD336 pKa = 3.87DD337 pKa = 5.23DD338 pKa = 3.93EE339 pKa = 5.05TVFFTFFEE347 pKa = 4.01QLRR350 pKa = 11.84PFNQAGEE357 pKa = 4.17IPEE360 pKa = 4.04VCANEE365 pKa = 4.2EE366 pKa = 4.26PGSIDD371 pKa = 5.36LFDD374 pKa = 4.76LLEE377 pKa = 4.08FTNEE381 pKa = 3.53NGIDD385 pKa = 4.22FIYY388 pKa = 11.1NDD390 pKa = 3.71DD391 pKa = 4.89AYY393 pKa = 11.26VKK395 pKa = 9.45WRR397 pKa = 11.84MVSGPSDD404 pKa = 3.58LPLSEE409 pKa = 4.32DD410 pKa = 3.32HH411 pKa = 6.92KK412 pKa = 11.2GTVQTLGAASGTYY425 pKa = 9.34VFEE428 pKa = 4.83FGVSPKK434 pKa = 9.94INCPGGEE441 pKa = 4.1AFCDD445 pKa = 3.57PFAAEE450 pKa = 4.56GEE452 pKa = 4.28EE453 pKa = 4.5GHH455 pKa = 7.0CEE457 pKa = 4.11FPCGVLTTQVTIRR470 pKa = 11.84VLEE473 pKa = 3.85FDD475 pKa = 4.07YY476 pKa = 11.32AGEE479 pKa = 4.25DD480 pKa = 3.26TDD482 pKa = 5.92GINICISEE490 pKa = 4.08NQIDD494 pKa = 4.43LRR496 pKa = 11.84SLLKK500 pKa = 10.35IEE502 pKa = 4.59GGNTIVDD509 pKa = 3.48TGVWTNEE516 pKa = 3.57AGEE519 pKa = 4.61EE520 pKa = 3.87IDD522 pKa = 3.49NTFVFPEE529 pKa = 4.04ISSPEE534 pKa = 3.91AFRR537 pKa = 11.84FTYY540 pKa = 9.55TATSDD545 pKa = 3.47MGCIDD550 pKa = 3.72AATLEE555 pKa = 4.54FTIHH559 pKa = 6.74PLPVAGEE566 pKa = 4.42DD567 pKa = 3.7GTASVCSDD575 pKa = 3.42NLTVTLFDD583 pKa = 4.19HH584 pKa = 7.45LEE586 pKa = 4.37GNPDD590 pKa = 3.25TTGIWTGPNGYY601 pKa = 8.05TSTDD605 pKa = 2.81HH606 pKa = 7.54LGRR609 pKa = 11.84FDD611 pKa = 4.82QNDD614 pKa = 3.52DD615 pKa = 3.62TLPILGAGRR624 pKa = 11.84FRR626 pKa = 11.84YY627 pKa = 7.99EE628 pKa = 3.76VPGNAGCLDD637 pKa = 3.71SDD639 pKa = 3.83ASFVDD644 pKa = 4.35VIIIEE649 pKa = 4.28PEE651 pKa = 3.87NIGNDD656 pKa = 3.44RR657 pKa = 11.84AEE659 pKa = 4.43TFCKK663 pKa = 9.62IDD665 pKa = 3.12GRR667 pKa = 11.84VNLFSLLDD675 pKa = 3.81RR676 pKa = 11.84NTVRR680 pKa = 11.84TGTFEE685 pKa = 4.32DD686 pKa = 4.08TDD688 pKa = 3.63DD689 pKa = 3.75TGALNSEE696 pKa = 4.19GVVEE700 pKa = 6.23FEE702 pKa = 4.15TLTNGIYY709 pKa = 9.99NFRR712 pKa = 11.84YY713 pKa = 9.65IISNTAPCEE722 pKa = 3.96EE723 pKa = 4.46SSLNVAIQIVDD734 pKa = 3.79LPEE737 pKa = 4.26PQVGDD742 pKa = 3.52QEE744 pKa = 4.92FCILDD749 pKa = 3.51AARR752 pKa = 11.84LEE754 pKa = 4.87DD755 pKa = 3.79IVVDD759 pKa = 3.33VLNYY763 pKa = 9.71NWYY766 pKa = 6.93EE767 pKa = 4.08TLEE770 pKa = 4.09GTIPIITNPILLDD783 pKa = 3.49EE784 pKa = 4.25QVFYY788 pKa = 10.74IANVDD793 pKa = 3.63ANNCEE798 pKa = 4.17SEE800 pKa = 4.2RR801 pKa = 11.84VEE803 pKa = 4.16VRR805 pKa = 11.84INILNIGEE813 pKa = 4.29RR814 pKa = 11.84SAQGEE819 pKa = 4.73LCTLDD824 pKa = 3.85FQDD827 pKa = 5.29GVSPNNDD834 pKa = 3.47VINDD838 pKa = 3.82TFDD841 pKa = 4.14LVIEE845 pKa = 4.26DD846 pKa = 4.4VYY848 pKa = 11.16NIPEE852 pKa = 4.5AFPDD856 pKa = 3.64FDD858 pKa = 4.72LKK860 pKa = 10.86IFNRR864 pKa = 11.84YY865 pKa = 8.99GSMVYY870 pKa = 10.02EE871 pKa = 4.52GNADD875 pKa = 3.55TEE877 pKa = 4.38EE878 pKa = 4.1FRR880 pKa = 11.84GEE882 pKa = 4.21SNISLRR888 pKa = 11.84LGDD891 pKa = 4.92DD892 pKa = 4.15LPSGTYY898 pKa = 9.9FYY900 pKa = 10.74IFTPNFEE907 pKa = 5.05NNFPIQGSFYY917 pKa = 10.87LSRR920 pKa = 5.17
Molecular weight: 101.33 kDa
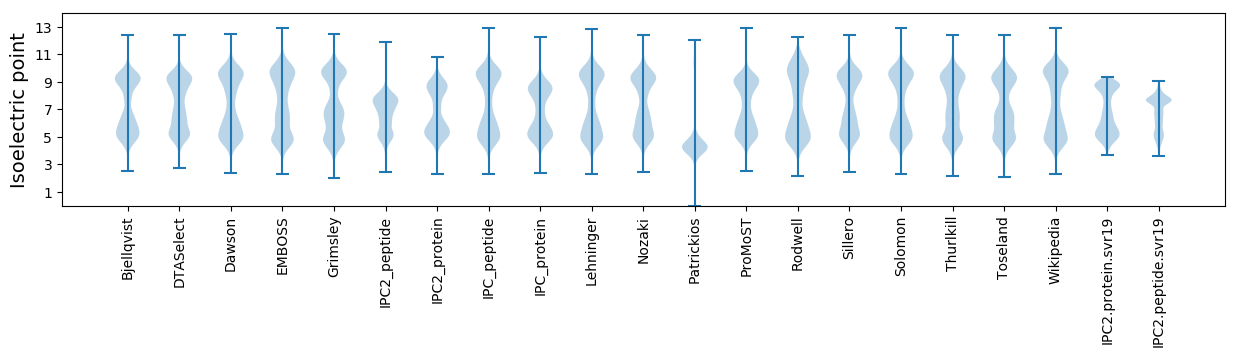
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7H8PNX5|A0A7H8PNX5_9FLAO M20/M25/M40 family metallo-hydrolase OS=Aquimarina sp. TRL1 OX=2736252 GN=HN014_14800 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.1VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.29KK37 pKa = 10.31GRR39 pKa = 11.84KK40 pKa = 7.92KK41 pKa = 10.69LSVSSEE47 pKa = 3.9LRR49 pKa = 11.84HH50 pKa = 5.43KK51 pKa = 10.29HH52 pKa = 5.04
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.1VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.29KK37 pKa = 10.31GRR39 pKa = 11.84KK40 pKa = 7.92KK41 pKa = 10.69LSVSSEE47 pKa = 3.9LRR49 pKa = 11.84HH50 pKa = 5.43KK51 pKa = 10.29HH52 pKa = 5.04
Molecular weight: 6.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1568201 |
15 |
9139 |
355.2 |
40.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.123 ± 0.042 | 0.791 ± 0.014 |
5.621 ± 0.122 | 6.605 ± 0.051 |
4.842 ± 0.047 | 6.452 ± 0.068 |
1.927 ± 0.025 | 8.208 ± 0.046 |
7.683 ± 0.089 | 8.952 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.984 ± 0.023 | 5.773 ± 0.044 |
3.425 ± 0.024 | 3.484 ± 0.027 |
3.534 ± 0.038 | 6.678 ± 0.034 |
6.524 ± 0.107 | 6.07 ± 0.038 |
1.044 ± 0.013 | 4.28 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
