
Capybara microvirus Cap1_SP_128
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.37
Get precalculated fractions of proteins
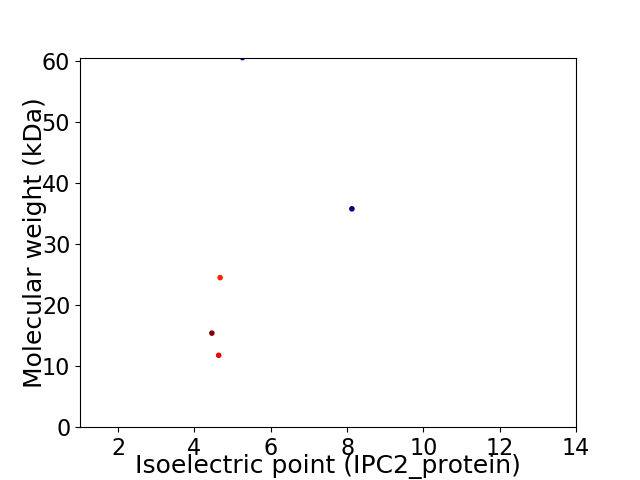
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W499|A0A4P8W499_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_128 OX=2585381 PE=4 SV=1
MM1 pKa = 7.61IYY3 pKa = 10.24FYY5 pKa = 11.55VVFYY9 pKa = 11.18KK10 pKa = 10.57KK11 pKa = 10.9GNDD14 pKa = 3.79YY15 pKa = 10.08QWSYY19 pKa = 11.65PFMHH23 pKa = 7.07PSIDD27 pKa = 3.19GVTNYY32 pKa = 9.88IIEE35 pKa = 4.11QLGVDD40 pKa = 3.48VEE42 pKa = 4.44IIKK45 pKa = 10.01VIRR48 pKa = 11.84LSCCDD53 pKa = 3.45MNNQVMIPYY62 pKa = 9.82RR63 pKa = 11.84EE64 pKa = 4.06EE65 pKa = 4.02IYY67 pKa = 11.01VDD69 pKa = 3.38EE70 pKa = 5.34KK71 pKa = 11.29LDD73 pKa = 3.9QEE75 pKa = 4.07FSKK78 pKa = 10.59IKK80 pKa = 10.21KK81 pKa = 9.46EE82 pKa = 4.65KK83 pKa = 9.3EE84 pKa = 3.5LQYY87 pKa = 11.05EE88 pKa = 4.03KK89 pKa = 11.29NEE91 pKa = 4.02NVEE94 pKa = 4.17SEE96 pKa = 4.38EE97 pKa = 4.01
MM1 pKa = 7.61IYY3 pKa = 10.24FYY5 pKa = 11.55VVFYY9 pKa = 11.18KK10 pKa = 10.57KK11 pKa = 10.9GNDD14 pKa = 3.79YY15 pKa = 10.08QWSYY19 pKa = 11.65PFMHH23 pKa = 7.07PSIDD27 pKa = 3.19GVTNYY32 pKa = 9.88IIEE35 pKa = 4.11QLGVDD40 pKa = 3.48VEE42 pKa = 4.44IIKK45 pKa = 10.01VIRR48 pKa = 11.84LSCCDD53 pKa = 3.45MNNQVMIPYY62 pKa = 9.82RR63 pKa = 11.84EE64 pKa = 4.06EE65 pKa = 4.02IYY67 pKa = 11.01VDD69 pKa = 3.38EE70 pKa = 5.34KK71 pKa = 11.29LDD73 pKa = 3.9QEE75 pKa = 4.07FSKK78 pKa = 10.59IKK80 pKa = 10.21KK81 pKa = 9.46EE82 pKa = 4.65KK83 pKa = 9.3EE84 pKa = 3.5LQYY87 pKa = 11.05EE88 pKa = 4.03KK89 pKa = 11.29NEE91 pKa = 4.02NVEE94 pKa = 4.17SEE96 pKa = 4.38EE97 pKa = 4.01
Molecular weight: 11.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7W7|A0A4P8W7W7_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_128 OX=2585381 PE=3 SV=1
MM1 pKa = 7.48EE2 pKa = 5.87CLNKK6 pKa = 10.21CHH8 pKa = 6.77VLISPNRR15 pKa = 11.84VIKK18 pKa = 10.66KK19 pKa = 10.38DD20 pKa = 3.28GTLGNRR26 pKa = 11.84VDD28 pKa = 4.55FFHH31 pKa = 7.11GLLSPVEE38 pKa = 4.14QSLLPAEE45 pKa = 4.36VRR47 pKa = 11.84YY48 pKa = 10.61EE49 pKa = 3.83EE50 pKa = 4.65VPCGKK55 pKa = 9.95CVNCKK60 pKa = 9.67RR61 pKa = 11.84SKK63 pKa = 10.7SYY65 pKa = 11.29AQFLRR70 pKa = 11.84LKK72 pKa = 9.42MEE74 pKa = 3.82QYY76 pKa = 10.66SYY78 pKa = 11.09KK79 pKa = 10.8SSDD82 pKa = 2.92ICFMTFTYY90 pKa = 10.95DD91 pKa = 3.53NYY93 pKa = 10.86HH94 pKa = 6.12LPKK97 pKa = 10.56DD98 pKa = 3.49RR99 pKa = 11.84LHH101 pKa = 6.84NYY103 pKa = 10.06RR104 pKa = 11.84DD105 pKa = 3.43FQLFMKK111 pKa = 10.21RR112 pKa = 11.84FRR114 pKa = 11.84KK115 pKa = 9.6HH116 pKa = 6.12FPADD120 pKa = 4.14DD121 pKa = 3.01IRR123 pKa = 11.84YY124 pKa = 8.25YY125 pKa = 10.95CVGEE129 pKa = 3.68YY130 pKa = 10.43GEE132 pKa = 4.9RR133 pKa = 11.84YY134 pKa = 9.2GRR136 pKa = 11.84PHH138 pKa = 5.32YY139 pKa = 10.34HH140 pKa = 6.35VILFGVDD147 pKa = 3.42LFSIADD153 pKa = 3.56YY154 pKa = 11.28HH155 pKa = 7.38DD156 pKa = 3.78VNEE159 pKa = 4.72NGNILYY165 pKa = 9.86YY166 pKa = 11.22SNVIDD171 pKa = 4.35KK172 pKa = 10.3LWNNGLVNFSEE183 pKa = 4.59VNDD186 pKa = 3.64NVFGYY191 pKa = 10.17VSQYY195 pKa = 9.8GVKK198 pKa = 10.37SLTSEE203 pKa = 4.27DD204 pKa = 3.67KK205 pKa = 10.81PLVRR209 pKa = 11.84SSRR212 pKa = 11.84RR213 pKa = 11.84GGLGYY218 pKa = 10.71RR219 pKa = 11.84FLDD222 pKa = 3.95DD223 pKa = 5.31NKK225 pKa = 11.28DD226 pKa = 3.59QIIKK230 pKa = 10.41NGYY233 pKa = 8.67ILLDD237 pKa = 3.69GKK239 pKa = 10.82KK240 pKa = 9.48YY241 pKa = 10.95NCDD244 pKa = 2.8KK245 pKa = 10.71YY246 pKa = 10.85VRR248 pKa = 11.84SFLGVSDD255 pKa = 4.29EE256 pKa = 3.88WTADD260 pKa = 3.39YY261 pKa = 10.63RR262 pKa = 11.84SKK264 pKa = 11.26LDD266 pKa = 3.48YY267 pKa = 10.35QHH269 pKa = 6.65SIKK272 pKa = 10.96VKK274 pKa = 7.48TTSLSDD280 pKa = 4.01DD281 pKa = 4.12YY282 pKa = 11.5INNILAQKK290 pKa = 9.9YY291 pKa = 7.23EE292 pKa = 3.82EE293 pKa = 4.13RR294 pKa = 11.84QRR296 pKa = 11.84AKK298 pKa = 10.94ALAKK302 pKa = 10.49KK303 pKa = 10.18LL304 pKa = 3.7
MM1 pKa = 7.48EE2 pKa = 5.87CLNKK6 pKa = 10.21CHH8 pKa = 6.77VLISPNRR15 pKa = 11.84VIKK18 pKa = 10.66KK19 pKa = 10.38DD20 pKa = 3.28GTLGNRR26 pKa = 11.84VDD28 pKa = 4.55FFHH31 pKa = 7.11GLLSPVEE38 pKa = 4.14QSLLPAEE45 pKa = 4.36VRR47 pKa = 11.84YY48 pKa = 10.61EE49 pKa = 3.83EE50 pKa = 4.65VPCGKK55 pKa = 9.95CVNCKK60 pKa = 9.67RR61 pKa = 11.84SKK63 pKa = 10.7SYY65 pKa = 11.29AQFLRR70 pKa = 11.84LKK72 pKa = 9.42MEE74 pKa = 3.82QYY76 pKa = 10.66SYY78 pKa = 11.09KK79 pKa = 10.8SSDD82 pKa = 2.92ICFMTFTYY90 pKa = 10.95DD91 pKa = 3.53NYY93 pKa = 10.86HH94 pKa = 6.12LPKK97 pKa = 10.56DD98 pKa = 3.49RR99 pKa = 11.84LHH101 pKa = 6.84NYY103 pKa = 10.06RR104 pKa = 11.84DD105 pKa = 3.43FQLFMKK111 pKa = 10.21RR112 pKa = 11.84FRR114 pKa = 11.84KK115 pKa = 9.6HH116 pKa = 6.12FPADD120 pKa = 4.14DD121 pKa = 3.01IRR123 pKa = 11.84YY124 pKa = 8.25YY125 pKa = 10.95CVGEE129 pKa = 3.68YY130 pKa = 10.43GEE132 pKa = 4.9RR133 pKa = 11.84YY134 pKa = 9.2GRR136 pKa = 11.84PHH138 pKa = 5.32YY139 pKa = 10.34HH140 pKa = 6.35VILFGVDD147 pKa = 3.42LFSIADD153 pKa = 3.56YY154 pKa = 11.28HH155 pKa = 7.38DD156 pKa = 3.78VNEE159 pKa = 4.72NGNILYY165 pKa = 9.86YY166 pKa = 11.22SNVIDD171 pKa = 4.35KK172 pKa = 10.3LWNNGLVNFSEE183 pKa = 4.59VNDD186 pKa = 3.64NVFGYY191 pKa = 10.17VSQYY195 pKa = 9.8GVKK198 pKa = 10.37SLTSEE203 pKa = 4.27DD204 pKa = 3.67KK205 pKa = 10.81PLVRR209 pKa = 11.84SSRR212 pKa = 11.84RR213 pKa = 11.84GGLGYY218 pKa = 10.71RR219 pKa = 11.84FLDD222 pKa = 3.95DD223 pKa = 5.31NKK225 pKa = 11.28DD226 pKa = 3.59QIIKK230 pKa = 10.41NGYY233 pKa = 8.67ILLDD237 pKa = 3.69GKK239 pKa = 10.82KK240 pKa = 9.48YY241 pKa = 10.95NCDD244 pKa = 2.8KK245 pKa = 10.71YY246 pKa = 10.85VRR248 pKa = 11.84SFLGVSDD255 pKa = 4.29EE256 pKa = 3.88WTADD260 pKa = 3.39YY261 pKa = 10.63RR262 pKa = 11.84SKK264 pKa = 11.26LDD266 pKa = 3.48YY267 pKa = 10.35QHH269 pKa = 6.65SIKK272 pKa = 10.96VKK274 pKa = 7.48TTSLSDD280 pKa = 4.01DD281 pKa = 4.12YY282 pKa = 11.5INNILAQKK290 pKa = 9.9YY291 pKa = 7.23EE292 pKa = 3.82EE293 pKa = 4.13RR294 pKa = 11.84QRR296 pKa = 11.84AKK298 pKa = 10.94ALAKK302 pKa = 10.49KK303 pKa = 10.18LL304 pKa = 3.7
Molecular weight: 35.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1304 |
97 |
539 |
260.8 |
29.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.525 ± 1.179 | 1.304 ± 0.44 |
7.439 ± 0.636 | 4.908 ± 0.926 |
4.601 ± 0.174 | 6.442 ± 0.465 |
2.147 ± 0.452 | 6.135 ± 0.743 |
5.598 ± 1.316 | 7.592 ± 0.824 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.147 ± 0.352 | 6.212 ± 0.737 |
4.601 ± 1.122 | 4.371 ± 0.403 |
4.371 ± 0.647 | 9.893 ± 1.296 |
4.525 ± 1.115 | 6.288 ± 0.593 |
1.227 ± 0.319 | 5.675 ± 0.969 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
