
Galinsoga mosaic virus (GaMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Gallantivirus
Average proteome isoelectric point is 7.9
Get precalculated fractions of proteins
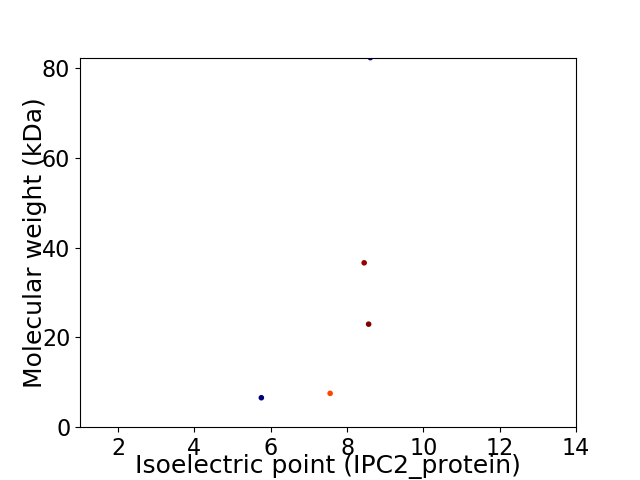
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|O12304|O12304_GAMV Capsid protein OS=Galinsoga mosaic virus OX=60714 PE=3 SV=1
MM1 pKa = 7.72KK2 pKa = 10.29YY3 pKa = 10.09CRR5 pKa = 11.84CSDD8 pKa = 3.73TAPTDD13 pKa = 3.96HH14 pKa = 6.62ITLLFVIFILSGLILSLCTNITSNNYY40 pKa = 9.08EE41 pKa = 3.76NHH43 pKa = 4.44TTEE46 pKa = 6.11NKK48 pKa = 7.9TQWITIGGQQQ58 pKa = 3.46
MM1 pKa = 7.72KK2 pKa = 10.29YY3 pKa = 10.09CRR5 pKa = 11.84CSDD8 pKa = 3.73TAPTDD13 pKa = 3.96HH14 pKa = 6.62ITLLFVIFILSGLILSLCTNITSNNYY40 pKa = 9.08EE41 pKa = 3.76NHH43 pKa = 4.44TTEE46 pKa = 6.11NKK48 pKa = 7.9TQWITIGGQQQ58 pKa = 3.46
Molecular weight: 6.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|O12302|O12302_GAMV Movement protein OS=Galinsoga mosaic virus OX=60714 PE=4 SV=1
MM1 pKa = 7.3GAISKK6 pKa = 9.91ISKK9 pKa = 9.44ISSFVVAGIVAYY21 pKa = 9.54KK22 pKa = 10.18VGKK25 pKa = 8.47ATYY28 pKa = 9.54NAFRR32 pKa = 11.84YY33 pKa = 9.63RR34 pKa = 11.84NQEE37 pKa = 4.06SITAARR43 pKa = 11.84DD44 pKa = 3.12LQAAVEE50 pKa = 4.17AVEE53 pKa = 4.15EE54 pKa = 4.14QPEE57 pKa = 4.19FGSVEE62 pKa = 4.08EE63 pKa = 4.67CLEE66 pKa = 4.14EE67 pKa = 4.45TPASQSEE74 pKa = 4.15RR75 pKa = 11.84TTIQNEE81 pKa = 3.62GDD83 pKa = 3.72LAKK86 pKa = 10.23MPVSRR91 pKa = 11.84KK92 pKa = 8.8RR93 pKa = 11.84RR94 pKa = 11.84IRR96 pKa = 11.84NRR98 pKa = 11.84KK99 pKa = 7.53HH100 pKa = 5.53GRR102 pKa = 11.84FVSYY106 pKa = 10.43LVNEE110 pKa = 4.73AKK112 pKa = 10.67AEE114 pKa = 4.02YY115 pKa = 9.95GLPKK119 pKa = 8.89PTEE122 pKa = 4.26AYY124 pKa = 10.2RR125 pKa = 11.84LMVGGFLNRR134 pKa = 11.84LCKK137 pKa = 9.86EE138 pKa = 3.83WGVVTSHH145 pKa = 5.5THH147 pKa = 3.93QAVSIALPLVFVPTKK162 pKa = 10.18FDD164 pKa = 3.44VMGKK168 pKa = 10.22AMCATTRR175 pKa = 11.84TNQLIGEE182 pKa = 4.32NSTDD186 pKa = 3.7QATGWFDD193 pKa = 3.3NVLGIGGGVGLRR205 pKa = 11.84FLGKK209 pKa = 8.09XGCLEE214 pKa = 3.78RR215 pKa = 11.84RR216 pKa = 11.84LGFSTTVSRR225 pKa = 11.84GSHH228 pKa = 6.85PDD230 pKa = 3.46LVVKK234 pKa = 10.26PSGRR238 pKa = 11.84PEE240 pKa = 3.84KK241 pKa = 10.24QRR243 pKa = 11.84QLSAIVDD250 pKa = 3.31SWGRR254 pKa = 11.84KK255 pKa = 8.08PLVFTITRR263 pKa = 11.84LANLRR268 pKa = 11.84RR269 pKa = 11.84GMIEE273 pKa = 3.3RR274 pKa = 11.84MYY276 pKa = 10.87YY277 pKa = 9.02VEE279 pKa = 4.12RR280 pKa = 11.84DD281 pKa = 3.55KK282 pKa = 11.82NLVEE286 pKa = 4.43PPAPSVDD293 pKa = 3.61FRR295 pKa = 11.84LPHH298 pKa = 6.49FSSAFKK304 pKa = 10.62KK305 pKa = 10.69VIGRR309 pKa = 11.84HH310 pKa = 4.38TPVTRR315 pKa = 11.84QAFVEE320 pKa = 5.42FYY322 pKa = 10.23RR323 pKa = 11.84GRR325 pKa = 11.84KK326 pKa = 6.22RR327 pKa = 11.84TIYY330 pKa = 9.69EE331 pKa = 3.93KK332 pKa = 10.72AVEE335 pKa = 4.36SLSAAPVTHH344 pKa = 7.16RR345 pKa = 11.84DD346 pKa = 3.17ARR348 pKa = 11.84LKK350 pKa = 10.3TFVKK354 pKa = 10.53AEE356 pKa = 3.98KK357 pKa = 10.25LVLSRR362 pKa = 11.84KK363 pKa = 9.02PDD365 pKa = 3.85PAPRR369 pKa = 11.84IIQPRR374 pKa = 11.84APRR377 pKa = 11.84YY378 pKa = 8.48NVEE381 pKa = 3.83LGVYY385 pKa = 9.83LRR387 pKa = 11.84PFEE390 pKa = 4.91HH391 pKa = 6.62KK392 pKa = 10.34AFRR395 pKa = 11.84AVEE398 pKa = 4.16RR399 pKa = 11.84VFGSPTIMKK408 pKa = 10.07GYY410 pKa = 7.65TAEE413 pKa = 4.13EE414 pKa = 3.87QASIARR420 pKa = 11.84EE421 pKa = 3.49KK422 pKa = 9.94WDD424 pKa = 3.65RR425 pKa = 11.84FSKK428 pKa = 10.42PVAIGLDD435 pKa = 3.26ASRR438 pKa = 11.84FDD440 pKa = 3.42QHH442 pKa = 8.35VSVAALKK449 pKa = 9.67WEE451 pKa = 4.21HH452 pKa = 6.55KK453 pKa = 10.07LYY455 pKa = 10.78HH456 pKa = 6.4HH457 pKa = 7.42AFNHH461 pKa = 6.58DD462 pKa = 3.36KK463 pKa = 10.22TLKK466 pKa = 10.64KK467 pKa = 10.04LLKK470 pKa = 9.06WQIHH474 pKa = 5.14NEE476 pKa = 3.9GTAFASDD483 pKa = 3.43GCIKK487 pKa = 10.48YY488 pKa = 10.38KK489 pKa = 9.74KK490 pKa = 8.03TGCRR494 pKa = 11.84MSGDD498 pKa = 3.79MNTSLGNCLIMCGMVYY514 pKa = 10.81DD515 pKa = 4.86LLVTHH520 pKa = 7.55DD521 pKa = 4.55IDD523 pKa = 6.31AEE525 pKa = 4.19LLNNGDD531 pKa = 4.17DD532 pKa = 3.85CVLICEE538 pKa = 4.46RR539 pKa = 11.84SEE541 pKa = 3.9EE542 pKa = 3.99AHH544 pKa = 6.44LLEE547 pKa = 5.83VIPQYY552 pKa = 10.48FRR554 pKa = 11.84QYY556 pKa = 9.4GFTMEE561 pKa = 4.15VEE563 pKa = 4.34KK564 pKa = 10.42PVHH567 pKa = 6.33IFEE570 pKa = 4.97KK571 pKa = 11.15LEE573 pKa = 3.99FCQTQPVYY581 pKa = 11.36VDD583 pKa = 3.83GKK585 pKa = 8.93WIMVRR590 pKa = 11.84KK591 pKa = 8.97PNPAMSKK598 pKa = 10.73DD599 pKa = 3.92LTCLLPIQNEE609 pKa = 4.31NNFKK613 pKa = 9.82EE614 pKa = 4.27WLSAVSEE621 pKa = 4.44CGLALTGGVPLWQNFYY637 pKa = 9.82QACNLGAARR646 pKa = 11.84TGFTKK651 pKa = 10.49SGLFTEE657 pKa = 4.98SGLAYY662 pKa = 10.23ASRR665 pKa = 11.84GLHH668 pKa = 5.58RR669 pKa = 11.84RR670 pKa = 11.84FSEE673 pKa = 4.14PSAEE677 pKa = 4.24TRR679 pKa = 11.84HH680 pKa = 5.3SFYY683 pKa = 10.76LAFGITPDD691 pKa = 3.7LQLALEE697 pKa = 4.78KK698 pKa = 10.75EE699 pKa = 4.31LDD701 pKa = 3.79SKK703 pKa = 11.32IMNEE707 pKa = 3.32WTMIPRR713 pKa = 11.84DD714 pKa = 3.61TRR716 pKa = 11.84QRR718 pKa = 11.84TYY720 pKa = 11.12YY721 pKa = 9.55STISNEE727 pKa = 3.97LTT729 pKa = 3.32
MM1 pKa = 7.3GAISKK6 pKa = 9.91ISKK9 pKa = 9.44ISSFVVAGIVAYY21 pKa = 9.54KK22 pKa = 10.18VGKK25 pKa = 8.47ATYY28 pKa = 9.54NAFRR32 pKa = 11.84YY33 pKa = 9.63RR34 pKa = 11.84NQEE37 pKa = 4.06SITAARR43 pKa = 11.84DD44 pKa = 3.12LQAAVEE50 pKa = 4.17AVEE53 pKa = 4.15EE54 pKa = 4.14QPEE57 pKa = 4.19FGSVEE62 pKa = 4.08EE63 pKa = 4.67CLEE66 pKa = 4.14EE67 pKa = 4.45TPASQSEE74 pKa = 4.15RR75 pKa = 11.84TTIQNEE81 pKa = 3.62GDD83 pKa = 3.72LAKK86 pKa = 10.23MPVSRR91 pKa = 11.84KK92 pKa = 8.8RR93 pKa = 11.84RR94 pKa = 11.84IRR96 pKa = 11.84NRR98 pKa = 11.84KK99 pKa = 7.53HH100 pKa = 5.53GRR102 pKa = 11.84FVSYY106 pKa = 10.43LVNEE110 pKa = 4.73AKK112 pKa = 10.67AEE114 pKa = 4.02YY115 pKa = 9.95GLPKK119 pKa = 8.89PTEE122 pKa = 4.26AYY124 pKa = 10.2RR125 pKa = 11.84LMVGGFLNRR134 pKa = 11.84LCKK137 pKa = 9.86EE138 pKa = 3.83WGVVTSHH145 pKa = 5.5THH147 pKa = 3.93QAVSIALPLVFVPTKK162 pKa = 10.18FDD164 pKa = 3.44VMGKK168 pKa = 10.22AMCATTRR175 pKa = 11.84TNQLIGEE182 pKa = 4.32NSTDD186 pKa = 3.7QATGWFDD193 pKa = 3.3NVLGIGGGVGLRR205 pKa = 11.84FLGKK209 pKa = 8.09XGCLEE214 pKa = 3.78RR215 pKa = 11.84RR216 pKa = 11.84LGFSTTVSRR225 pKa = 11.84GSHH228 pKa = 6.85PDD230 pKa = 3.46LVVKK234 pKa = 10.26PSGRR238 pKa = 11.84PEE240 pKa = 3.84KK241 pKa = 10.24QRR243 pKa = 11.84QLSAIVDD250 pKa = 3.31SWGRR254 pKa = 11.84KK255 pKa = 8.08PLVFTITRR263 pKa = 11.84LANLRR268 pKa = 11.84RR269 pKa = 11.84GMIEE273 pKa = 3.3RR274 pKa = 11.84MYY276 pKa = 10.87YY277 pKa = 9.02VEE279 pKa = 4.12RR280 pKa = 11.84DD281 pKa = 3.55KK282 pKa = 11.82NLVEE286 pKa = 4.43PPAPSVDD293 pKa = 3.61FRR295 pKa = 11.84LPHH298 pKa = 6.49FSSAFKK304 pKa = 10.62KK305 pKa = 10.69VIGRR309 pKa = 11.84HH310 pKa = 4.38TPVTRR315 pKa = 11.84QAFVEE320 pKa = 5.42FYY322 pKa = 10.23RR323 pKa = 11.84GRR325 pKa = 11.84KK326 pKa = 6.22RR327 pKa = 11.84TIYY330 pKa = 9.69EE331 pKa = 3.93KK332 pKa = 10.72AVEE335 pKa = 4.36SLSAAPVTHH344 pKa = 7.16RR345 pKa = 11.84DD346 pKa = 3.17ARR348 pKa = 11.84LKK350 pKa = 10.3TFVKK354 pKa = 10.53AEE356 pKa = 3.98KK357 pKa = 10.25LVLSRR362 pKa = 11.84KK363 pKa = 9.02PDD365 pKa = 3.85PAPRR369 pKa = 11.84IIQPRR374 pKa = 11.84APRR377 pKa = 11.84YY378 pKa = 8.48NVEE381 pKa = 3.83LGVYY385 pKa = 9.83LRR387 pKa = 11.84PFEE390 pKa = 4.91HH391 pKa = 6.62KK392 pKa = 10.34AFRR395 pKa = 11.84AVEE398 pKa = 4.16RR399 pKa = 11.84VFGSPTIMKK408 pKa = 10.07GYY410 pKa = 7.65TAEE413 pKa = 4.13EE414 pKa = 3.87QASIARR420 pKa = 11.84EE421 pKa = 3.49KK422 pKa = 9.94WDD424 pKa = 3.65RR425 pKa = 11.84FSKK428 pKa = 10.42PVAIGLDD435 pKa = 3.26ASRR438 pKa = 11.84FDD440 pKa = 3.42QHH442 pKa = 8.35VSVAALKK449 pKa = 9.67WEE451 pKa = 4.21HH452 pKa = 6.55KK453 pKa = 10.07LYY455 pKa = 10.78HH456 pKa = 6.4HH457 pKa = 7.42AFNHH461 pKa = 6.58DD462 pKa = 3.36KK463 pKa = 10.22TLKK466 pKa = 10.64KK467 pKa = 10.04LLKK470 pKa = 9.06WQIHH474 pKa = 5.14NEE476 pKa = 3.9GTAFASDD483 pKa = 3.43GCIKK487 pKa = 10.48YY488 pKa = 10.38KK489 pKa = 9.74KK490 pKa = 8.03TGCRR494 pKa = 11.84MSGDD498 pKa = 3.79MNTSLGNCLIMCGMVYY514 pKa = 10.81DD515 pKa = 4.86LLVTHH520 pKa = 7.55DD521 pKa = 4.55IDD523 pKa = 6.31AEE525 pKa = 4.19LLNNGDD531 pKa = 4.17DD532 pKa = 3.85CVLICEE538 pKa = 4.46RR539 pKa = 11.84SEE541 pKa = 3.9EE542 pKa = 3.99AHH544 pKa = 6.44LLEE547 pKa = 5.83VIPQYY552 pKa = 10.48FRR554 pKa = 11.84QYY556 pKa = 9.4GFTMEE561 pKa = 4.15VEE563 pKa = 4.34KK564 pKa = 10.42PVHH567 pKa = 6.33IFEE570 pKa = 4.97KK571 pKa = 11.15LEE573 pKa = 3.99FCQTQPVYY581 pKa = 11.36VDD583 pKa = 3.83GKK585 pKa = 8.93WIMVRR590 pKa = 11.84KK591 pKa = 8.97PNPAMSKK598 pKa = 10.73DD599 pKa = 3.92LTCLLPIQNEE609 pKa = 4.31NNFKK613 pKa = 9.82EE614 pKa = 4.27WLSAVSEE621 pKa = 4.44CGLALTGGVPLWQNFYY637 pKa = 9.82QACNLGAARR646 pKa = 11.84TGFTKK651 pKa = 10.49SGLFTEE657 pKa = 4.98SGLAYY662 pKa = 10.23ASRR665 pKa = 11.84GLHH668 pKa = 5.58RR669 pKa = 11.84RR670 pKa = 11.84FSEE673 pKa = 4.14PSAEE677 pKa = 4.24TRR679 pKa = 11.84HH680 pKa = 5.3SFYY683 pKa = 10.76LAFGITPDD691 pKa = 3.7LQLALEE697 pKa = 4.78KK698 pKa = 10.75EE699 pKa = 4.31LDD701 pKa = 3.79SKK703 pKa = 11.32IMNEE707 pKa = 3.32WTMIPRR713 pKa = 11.84DD714 pKa = 3.61TRR716 pKa = 11.84QRR718 pKa = 11.84TYY720 pKa = 11.12YY721 pKa = 9.55STISNEE727 pKa = 3.97LTT729 pKa = 3.32
Molecular weight: 82.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1405 |
58 |
729 |
281.0 |
31.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.473 ± 0.932 | 1.637 ± 0.398 |
3.63 ± 0.291 | 5.694 ± 1.314 |
4.626 ± 0.293 | 7.758 ± 0.787 |
2.206 ± 0.36 | 4.555 ± 0.709 |
5.338 ± 0.94 | 7.829 ± 0.896 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.993 ± 0.504 | 5.125 ± 1.551 |
4.555 ± 0.536 | 4.342 ± 0.851 |
6.406 ± 0.926 | 7.331 ± 0.936 |
7.829 ± 1.565 | 7.26 ± 0.624 |
1.139 ± 0.184 | 3.203 ± 0.22 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
