
Chloris striate mosaic virus (CSMV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus
Average proteome isoelectric point is 7.44
Get precalculated fractions of proteins
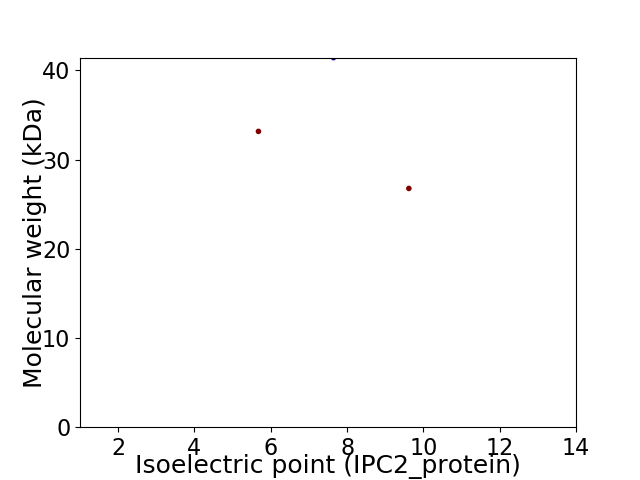
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
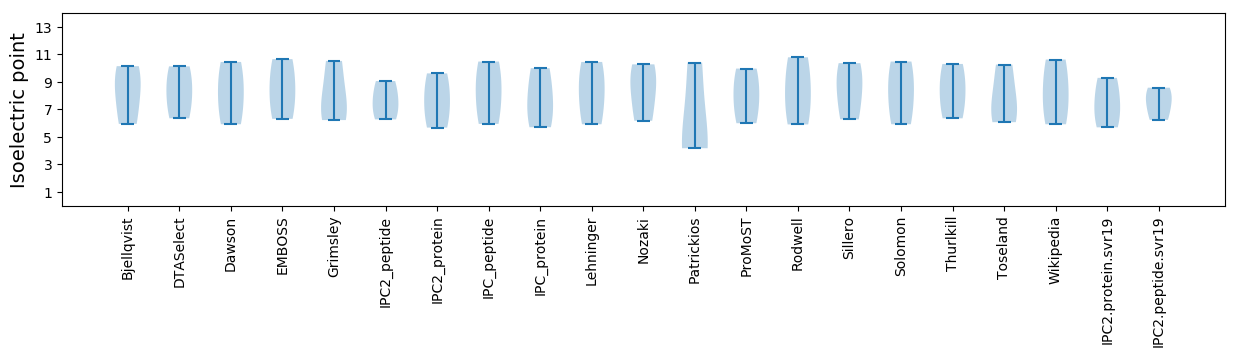
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P18921|REPA_CSMV Replication-associated protein A OS=Chloris striate mosaic virus OX=10820 GN=C1 PE=3 SV=1
MM1 pKa = 7.48SSLPVSEE8 pKa = 4.93SEE10 pKa = 4.53GEE12 pKa = 4.15GSGTSVQVPSRR23 pKa = 11.84GGQVTPGEE31 pKa = 4.43KK32 pKa = 10.33AFSLRR37 pKa = 11.84TKK39 pKa = 10.36HH40 pKa = 6.58VFLTYY45 pKa = 9.29PRR47 pKa = 11.84CPISPEE53 pKa = 3.63EE54 pKa = 4.19AGQKK58 pKa = 9.55IADD61 pKa = 3.89RR62 pKa = 11.84LKK64 pKa = 10.81NKK66 pKa = 8.98KK67 pKa = 9.79CNYY70 pKa = 9.36IYY72 pKa = 10.41ISRR75 pKa = 11.84EE76 pKa = 3.42FHH78 pKa = 7.42ADD80 pKa = 3.65GEE82 pKa = 4.57PHH84 pKa = 6.35LHH86 pKa = 6.94AFVQLEE92 pKa = 4.18ANFRR96 pKa = 11.84TTSPKK101 pKa = 10.57YY102 pKa = 9.07FDD104 pKa = 4.96LDD106 pKa = 3.54EE107 pKa = 4.3FHH109 pKa = 7.51PNIQAARR116 pKa = 11.84QPASTLKK123 pKa = 9.73YY124 pKa = 10.1CMKK127 pKa = 10.23HH128 pKa = 6.39PEE130 pKa = 4.17SSWEE134 pKa = 3.67FGKK137 pKa = 10.47FLKK140 pKa = 10.6PKK142 pKa = 9.64VNRR145 pKa = 11.84SPTQSASRR153 pKa = 11.84DD154 pKa = 3.37KK155 pKa = 11.08TMKK158 pKa = 10.43QIMANATSRR167 pKa = 11.84DD168 pKa = 3.71EE169 pKa = 4.09YY170 pKa = 11.18LSMVRR175 pKa = 11.84KK176 pKa = 10.02SFPFEE181 pKa = 3.44WAVRR185 pKa = 11.84LQQFQYY191 pKa = 10.6SANALFPDD199 pKa = 4.65PPQTYY204 pKa = 8.8SAPYY208 pKa = 9.77ASRR211 pKa = 11.84DD212 pKa = 3.67MSDD215 pKa = 3.35HH216 pKa = 6.14PVIGEE221 pKa = 3.86WLQQEE226 pKa = 5.58LYY228 pKa = 9.55TVSPQALSLHH238 pKa = 6.3AGISEE243 pKa = 4.0EE244 pKa = 3.98QARR247 pKa = 11.84IDD249 pKa = 4.02LQWMSDD255 pKa = 3.57LTRR258 pKa = 11.84SGALEE263 pKa = 4.49SGDD266 pKa = 3.86EE267 pKa = 4.29ACTSVGQQEE276 pKa = 4.49LEE278 pKa = 3.94RR279 pKa = 11.84LLGPEE284 pKa = 4.04VLEE287 pKa = 5.53LITTGSTQQ295 pKa = 2.91
MM1 pKa = 7.48SSLPVSEE8 pKa = 4.93SEE10 pKa = 4.53GEE12 pKa = 4.15GSGTSVQVPSRR23 pKa = 11.84GGQVTPGEE31 pKa = 4.43KK32 pKa = 10.33AFSLRR37 pKa = 11.84TKK39 pKa = 10.36HH40 pKa = 6.58VFLTYY45 pKa = 9.29PRR47 pKa = 11.84CPISPEE53 pKa = 3.63EE54 pKa = 4.19AGQKK58 pKa = 9.55IADD61 pKa = 3.89RR62 pKa = 11.84LKK64 pKa = 10.81NKK66 pKa = 8.98KK67 pKa = 9.79CNYY70 pKa = 9.36IYY72 pKa = 10.41ISRR75 pKa = 11.84EE76 pKa = 3.42FHH78 pKa = 7.42ADD80 pKa = 3.65GEE82 pKa = 4.57PHH84 pKa = 6.35LHH86 pKa = 6.94AFVQLEE92 pKa = 4.18ANFRR96 pKa = 11.84TTSPKK101 pKa = 10.57YY102 pKa = 9.07FDD104 pKa = 4.96LDD106 pKa = 3.54EE107 pKa = 4.3FHH109 pKa = 7.51PNIQAARR116 pKa = 11.84QPASTLKK123 pKa = 9.73YY124 pKa = 10.1CMKK127 pKa = 10.23HH128 pKa = 6.39PEE130 pKa = 4.17SSWEE134 pKa = 3.67FGKK137 pKa = 10.47FLKK140 pKa = 10.6PKK142 pKa = 9.64VNRR145 pKa = 11.84SPTQSASRR153 pKa = 11.84DD154 pKa = 3.37KK155 pKa = 11.08TMKK158 pKa = 10.43QIMANATSRR167 pKa = 11.84DD168 pKa = 3.71EE169 pKa = 4.09YY170 pKa = 11.18LSMVRR175 pKa = 11.84KK176 pKa = 10.02SFPFEE181 pKa = 3.44WAVRR185 pKa = 11.84LQQFQYY191 pKa = 10.6SANALFPDD199 pKa = 4.65PPQTYY204 pKa = 8.8SAPYY208 pKa = 9.77ASRR211 pKa = 11.84DD212 pKa = 3.67MSDD215 pKa = 3.35HH216 pKa = 6.14PVIGEE221 pKa = 3.86WLQQEE226 pKa = 5.58LYY228 pKa = 9.55TVSPQALSLHH238 pKa = 6.3AGISEE243 pKa = 4.0EE244 pKa = 3.98QARR247 pKa = 11.84IDD249 pKa = 4.02LQWMSDD255 pKa = 3.57LTRR258 pKa = 11.84SGALEE263 pKa = 4.49SGDD266 pKa = 3.86EE267 pKa = 4.29ACTSVGQQEE276 pKa = 4.49LEE278 pKa = 3.94RR279 pKa = 11.84LLGPEE284 pKa = 4.04VLEE287 pKa = 5.53LITTGSTQQ295 pKa = 2.91
Molecular weight: 33.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P18919|REP_CSMV Replication-associated protein OS=Chloris striate mosaic virus OX=10820 GN=C1/C2 PE=3 SV=2
MM1 pKa = 7.48SPASSWKK8 pKa = 9.62RR9 pKa = 11.84KK10 pKa = 9.48RR11 pKa = 11.84PSSSSAQASKK21 pKa = 10.56KK22 pKa = 9.43RR23 pKa = 11.84RR24 pKa = 11.84VYY26 pKa = 10.13RR27 pKa = 11.84PAVSRR32 pKa = 11.84SLARR36 pKa = 11.84RR37 pKa = 11.84EE38 pKa = 3.83PLQVQDD44 pKa = 5.05FVWDD48 pKa = 3.35TDD50 pKa = 3.64VAFNRR55 pKa = 11.84GGGCYY60 pKa = 10.38LLTSYY65 pKa = 11.4ARR67 pKa = 11.84GSAEE71 pKa = 4.04NQRR74 pKa = 11.84KK75 pKa = 5.73TAEE78 pKa = 4.12TITYY82 pKa = 9.28KK83 pKa = 10.76VAVNLGCAISGTMQQYY99 pKa = 10.37CISSRR104 pKa = 11.84PVCWIVYY111 pKa = 9.2DD112 pKa = 4.12AAPTGSAVTPKK123 pKa = 10.6DD124 pKa = 2.86IFGYY128 pKa = 10.02PEE130 pKa = 4.68GLVNWPTTWKK140 pKa = 9.23VARR143 pKa = 11.84AVSHH147 pKa = 6.15RR148 pKa = 11.84FIVKK152 pKa = 9.7RR153 pKa = 11.84RR154 pKa = 11.84WVFTMEE160 pKa = 4.52SNGSRR165 pKa = 11.84FDD167 pKa = 3.73RR168 pKa = 11.84DD169 pKa = 3.8YY170 pKa = 11.11TNLPAAIPQSLPVLNKK186 pKa = 9.32FAKK189 pKa = 9.26QLGVRR194 pKa = 11.84TEE196 pKa = 3.78WKK198 pKa = 9.77NAEE201 pKa = 4.27GGDD204 pKa = 3.68FGDD207 pKa = 3.81IKK209 pKa = 11.1SGALYY214 pKa = 10.76LVMAPANGAVFVARR228 pKa = 11.84GNVRR232 pKa = 11.84VYY234 pKa = 10.13FKK236 pKa = 11.12SVGNQQ241 pKa = 2.8
MM1 pKa = 7.48SPASSWKK8 pKa = 9.62RR9 pKa = 11.84KK10 pKa = 9.48RR11 pKa = 11.84PSSSSAQASKK21 pKa = 10.56KK22 pKa = 9.43RR23 pKa = 11.84RR24 pKa = 11.84VYY26 pKa = 10.13RR27 pKa = 11.84PAVSRR32 pKa = 11.84SLARR36 pKa = 11.84RR37 pKa = 11.84EE38 pKa = 3.83PLQVQDD44 pKa = 5.05FVWDD48 pKa = 3.35TDD50 pKa = 3.64VAFNRR55 pKa = 11.84GGGCYY60 pKa = 10.38LLTSYY65 pKa = 11.4ARR67 pKa = 11.84GSAEE71 pKa = 4.04NQRR74 pKa = 11.84KK75 pKa = 5.73TAEE78 pKa = 4.12TITYY82 pKa = 9.28KK83 pKa = 10.76VAVNLGCAISGTMQQYY99 pKa = 10.37CISSRR104 pKa = 11.84PVCWIVYY111 pKa = 9.2DD112 pKa = 4.12AAPTGSAVTPKK123 pKa = 10.6DD124 pKa = 2.86IFGYY128 pKa = 10.02PEE130 pKa = 4.68GLVNWPTTWKK140 pKa = 9.23VARR143 pKa = 11.84AVSHH147 pKa = 6.15RR148 pKa = 11.84FIVKK152 pKa = 9.7RR153 pKa = 11.84RR154 pKa = 11.84WVFTMEE160 pKa = 4.52SNGSRR165 pKa = 11.84FDD167 pKa = 3.73RR168 pKa = 11.84DD169 pKa = 3.8YY170 pKa = 11.11TNLPAAIPQSLPVLNKK186 pKa = 9.32FAKK189 pKa = 9.26QLGVRR194 pKa = 11.84TEE196 pKa = 3.78WKK198 pKa = 9.77NAEE201 pKa = 4.27GGDD204 pKa = 3.68FGDD207 pKa = 3.81IKK209 pKa = 11.1SGALYY214 pKa = 10.76LVMAPANGAVFVARR228 pKa = 11.84GNVRR232 pKa = 11.84VYY234 pKa = 10.13FKK236 pKa = 11.12SVGNQQ241 pKa = 2.8
Molecular weight: 26.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
897 |
241 |
361 |
299.0 |
33.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.469 ± 1.082 | 1.672 ± 0.143 |
4.013 ± 0.098 | 5.909 ± 1.218 |
4.682 ± 0.268 | 6.02 ± 0.514 |
2.341 ± 0.68 | 3.79 ± 0.291 |
5.686 ± 0.257 | 6.577 ± 0.746 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.007 ± 0.17 | 3.567 ± 0.536 |
6.912 ± 0.542 | 5.574 ± 0.713 |
6.243 ± 0.867 | 9.699 ± 0.484 |
5.463 ± 0.128 | 6.132 ± 1.242 |
2.341 ± 0.418 | 3.902 ± 0.216 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
