
Yonghaparkia sp. Root332
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Yonghaparkia; unclassified Yonghaparkia
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
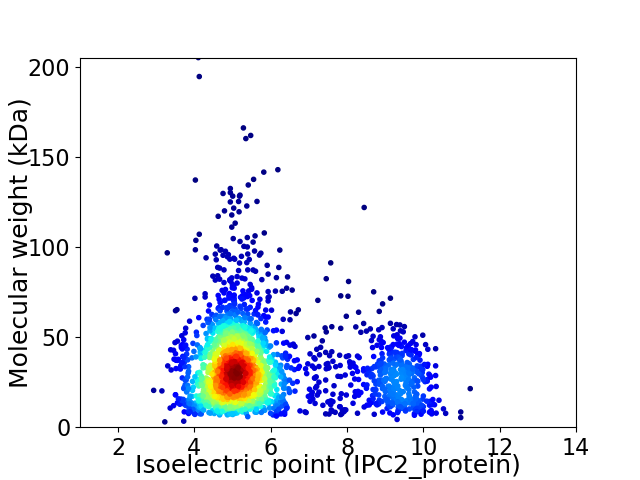
Virtual 2D-PAGE plot for 2389 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
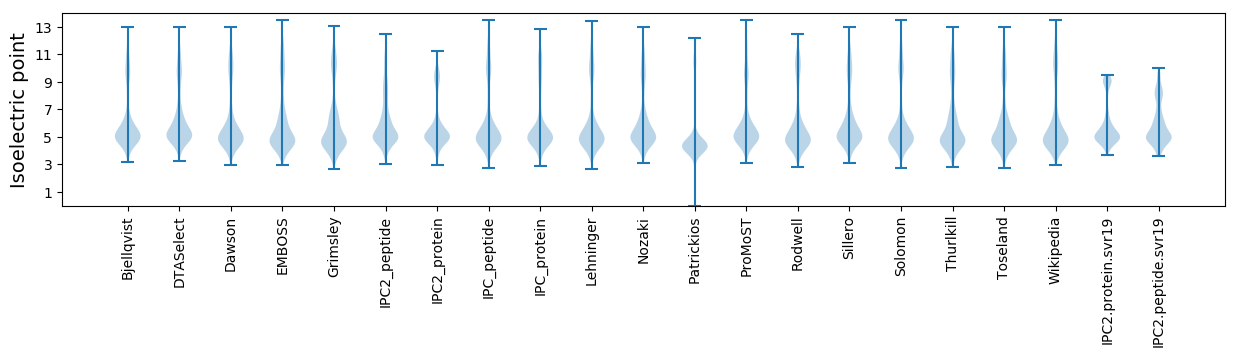
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q6R9I8|A0A0Q6R9I8_9MICO 50S ribosomal protein L18 OS=Yonghaparkia sp. Root332 OX=1736516 GN=rplR PE=3 SV=1
MM1 pKa = 7.49RR2 pKa = 11.84TRR4 pKa = 11.84TRR6 pKa = 11.84LSAVALPLVAVLALSGCVPTWLSGLTGQDD35 pKa = 2.68TSTPTGEE42 pKa = 4.11EE43 pKa = 3.8VEE45 pKa = 4.58AALEE49 pKa = 4.09PYY51 pKa = 9.58YY52 pKa = 10.61RR53 pKa = 11.84QALVWEE59 pKa = 4.49RR60 pKa = 11.84CGGGAQCATAIAPLDD75 pKa = 3.59WDD77 pKa = 4.55DD78 pKa = 5.09PGAGDD83 pKa = 5.49DD84 pKa = 4.4IEE86 pKa = 4.53LALVRR91 pKa = 11.84YY92 pKa = 8.72RR93 pKa = 11.84ATDD96 pKa = 3.33EE97 pKa = 4.27AQGSLFVNPGGPGASGFDD115 pKa = 3.64FVRR118 pKa = 11.84DD119 pKa = 3.69SVDD122 pKa = 3.73FAVSEE127 pKa = 4.04RR128 pKa = 11.84LRR130 pKa = 11.84ANFDD134 pKa = 3.73VIGWDD139 pKa = 3.26PRR141 pKa = 11.84GVGRR145 pKa = 11.84SSAVTCFTDD154 pKa = 3.73PADD157 pKa = 3.68MDD159 pKa = 3.75EE160 pKa = 5.21FIYY163 pKa = 9.98GVPEE167 pKa = 4.3AEE169 pKa = 4.36PEE171 pKa = 4.18TPEE174 pKa = 3.53WVAEE178 pKa = 4.26VEE180 pKa = 4.4RR181 pKa = 11.84SAQRR185 pKa = 11.84FVDD188 pKa = 3.91ACAEE192 pKa = 4.1NTGPLLEE199 pKa = 5.3HH200 pKa = 7.03VDD202 pKa = 3.38TDD204 pKa = 3.78STVRR208 pKa = 11.84DD209 pKa = 3.69LDD211 pKa = 3.38MLRR214 pKa = 11.84AVVGDD219 pKa = 3.34EE220 pKa = 3.46ALNYY224 pKa = 10.0FGYY227 pKa = 10.43SYY229 pKa = 9.97GTEE232 pKa = 3.36IGARR236 pKa = 11.84YY237 pKa = 9.8ADD239 pKa = 3.94RR240 pKa = 11.84FPDD243 pKa = 3.26RR244 pKa = 11.84VGRR247 pKa = 11.84LVLDD251 pKa = 4.61GATDD255 pKa = 3.48PTTTQFEE262 pKa = 4.64VVLAQSIGFEE272 pKa = 4.16AALTTYY278 pKa = 9.33LTEE281 pKa = 4.3CSAAASCPFPTDD293 pKa = 2.95TTAALAIVEE302 pKa = 3.98QLYY305 pKa = 9.39QQLEE309 pKa = 4.16ADD311 pKa = 5.59PIPAADD317 pKa = 3.56GRR319 pKa = 11.84LFTDD323 pKa = 3.44SVLDD327 pKa = 3.59IAVATALYY335 pKa = 10.68DD336 pKa = 3.6EE337 pKa = 5.37ASWEE341 pKa = 4.16FLSQMFTEE349 pKa = 4.53LRR351 pKa = 11.84TGVVEE356 pKa = 4.3TGFLLADD363 pKa = 4.25FYY365 pKa = 11.6YY366 pKa = 10.74GRR368 pKa = 11.84EE369 pKa = 3.75NGEE372 pKa = 3.99YY373 pKa = 10.41VDD375 pKa = 4.47NSLEE379 pKa = 3.8AFIAINCLDD388 pKa = 3.98YY389 pKa = 10.84PVEE392 pKa = 4.74RR393 pKa = 11.84DD394 pKa = 3.14PEE396 pKa = 4.53TIVEE400 pKa = 4.08QNAAIAEE407 pKa = 4.28AAPVTSDD414 pKa = 3.27PSILGDD420 pKa = 3.41VVCQRR425 pKa = 11.84WPFAFEE431 pKa = 4.08GQLGPVSAEE440 pKa = 3.71GAPPILVVGTTGDD453 pKa = 3.54PATPYY458 pKa = 10.23VWAEE462 pKa = 3.85ALASQLDD469 pKa = 3.95SGVLLTYY476 pKa = 10.59DD477 pKa = 3.85GEE479 pKa = 4.33GHH481 pKa = 6.43IAYY484 pKa = 10.04DD485 pKa = 4.62EE486 pKa = 4.59GDD488 pKa = 3.45PCINEE493 pKa = 4.32LVDD496 pKa = 4.67DD497 pKa = 4.38YY498 pKa = 11.18FVTGAVPARR507 pKa = 11.84DD508 pKa = 3.84PVCAGG513 pKa = 3.26
MM1 pKa = 7.49RR2 pKa = 11.84TRR4 pKa = 11.84TRR6 pKa = 11.84LSAVALPLVAVLALSGCVPTWLSGLTGQDD35 pKa = 2.68TSTPTGEE42 pKa = 4.11EE43 pKa = 3.8VEE45 pKa = 4.58AALEE49 pKa = 4.09PYY51 pKa = 9.58YY52 pKa = 10.61RR53 pKa = 11.84QALVWEE59 pKa = 4.49RR60 pKa = 11.84CGGGAQCATAIAPLDD75 pKa = 3.59WDD77 pKa = 4.55DD78 pKa = 5.09PGAGDD83 pKa = 5.49DD84 pKa = 4.4IEE86 pKa = 4.53LALVRR91 pKa = 11.84YY92 pKa = 8.72RR93 pKa = 11.84ATDD96 pKa = 3.33EE97 pKa = 4.27AQGSLFVNPGGPGASGFDD115 pKa = 3.64FVRR118 pKa = 11.84DD119 pKa = 3.69SVDD122 pKa = 3.73FAVSEE127 pKa = 4.04RR128 pKa = 11.84LRR130 pKa = 11.84ANFDD134 pKa = 3.73VIGWDD139 pKa = 3.26PRR141 pKa = 11.84GVGRR145 pKa = 11.84SSAVTCFTDD154 pKa = 3.73PADD157 pKa = 3.68MDD159 pKa = 3.75EE160 pKa = 5.21FIYY163 pKa = 9.98GVPEE167 pKa = 4.3AEE169 pKa = 4.36PEE171 pKa = 4.18TPEE174 pKa = 3.53WVAEE178 pKa = 4.26VEE180 pKa = 4.4RR181 pKa = 11.84SAQRR185 pKa = 11.84FVDD188 pKa = 3.91ACAEE192 pKa = 4.1NTGPLLEE199 pKa = 5.3HH200 pKa = 7.03VDD202 pKa = 3.38TDD204 pKa = 3.78STVRR208 pKa = 11.84DD209 pKa = 3.69LDD211 pKa = 3.38MLRR214 pKa = 11.84AVVGDD219 pKa = 3.34EE220 pKa = 3.46ALNYY224 pKa = 10.0FGYY227 pKa = 10.43SYY229 pKa = 9.97GTEE232 pKa = 3.36IGARR236 pKa = 11.84YY237 pKa = 9.8ADD239 pKa = 3.94RR240 pKa = 11.84FPDD243 pKa = 3.26RR244 pKa = 11.84VGRR247 pKa = 11.84LVLDD251 pKa = 4.61GATDD255 pKa = 3.48PTTTQFEE262 pKa = 4.64VVLAQSIGFEE272 pKa = 4.16AALTTYY278 pKa = 9.33LTEE281 pKa = 4.3CSAAASCPFPTDD293 pKa = 2.95TTAALAIVEE302 pKa = 3.98QLYY305 pKa = 9.39QQLEE309 pKa = 4.16ADD311 pKa = 5.59PIPAADD317 pKa = 3.56GRR319 pKa = 11.84LFTDD323 pKa = 3.44SVLDD327 pKa = 3.59IAVATALYY335 pKa = 10.68DD336 pKa = 3.6EE337 pKa = 5.37ASWEE341 pKa = 4.16FLSQMFTEE349 pKa = 4.53LRR351 pKa = 11.84TGVVEE356 pKa = 4.3TGFLLADD363 pKa = 4.25FYY365 pKa = 11.6YY366 pKa = 10.74GRR368 pKa = 11.84EE369 pKa = 3.75NGEE372 pKa = 3.99YY373 pKa = 10.41VDD375 pKa = 4.47NSLEE379 pKa = 3.8AFIAINCLDD388 pKa = 3.98YY389 pKa = 10.84PVEE392 pKa = 4.74RR393 pKa = 11.84DD394 pKa = 3.14PEE396 pKa = 4.53TIVEE400 pKa = 4.08QNAAIAEE407 pKa = 4.28AAPVTSDD414 pKa = 3.27PSILGDD420 pKa = 3.41VVCQRR425 pKa = 11.84WPFAFEE431 pKa = 4.08GQLGPVSAEE440 pKa = 3.71GAPPILVVGTTGDD453 pKa = 3.54PATPYY458 pKa = 10.23VWAEE462 pKa = 3.85ALASQLDD469 pKa = 3.95SGVLLTYY476 pKa = 10.59DD477 pKa = 3.85GEE479 pKa = 4.33GHH481 pKa = 6.43IAYY484 pKa = 10.04DD485 pKa = 4.62EE486 pKa = 4.59GDD488 pKa = 3.45PCINEE493 pKa = 4.32LVDD496 pKa = 4.67DD497 pKa = 4.38YY498 pKa = 11.18FVTGAVPARR507 pKa = 11.84DD508 pKa = 3.84PVCAGG513 pKa = 3.26
Molecular weight: 55.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q6RN09|A0A0Q6RN09_9MICO Malto-oligosyltrehalose trehalohydrolase OS=Yonghaparkia sp. Root332 OX=1736516 GN=ASC54_02485 PE=3 SV=1
MM1 pKa = 7.07TRR3 pKa = 11.84SLHH6 pKa = 5.25FVRR9 pKa = 11.84SARR12 pKa = 11.84PHH14 pKa = 6.65ALLACAGAPSRR25 pKa = 11.84AVAPLGPRR33 pKa = 11.84TARR36 pKa = 11.84RR37 pKa = 11.84VIAPRR42 pKa = 11.84TVVARR47 pKa = 11.84RR48 pKa = 11.84IPVALPGAVAARR60 pKa = 11.84GAVLLPRR67 pKa = 11.84AVASRR72 pKa = 11.84RR73 pKa = 11.84IAVTARR79 pKa = 11.84TIGAVRR85 pKa = 11.84PCPARR90 pKa = 11.84SIAVTLPRR98 pKa = 11.84LIATRR103 pKa = 11.84SLAIPLPRR111 pKa = 11.84PVAARR116 pKa = 11.84RR117 pKa = 11.84AVLLPRR123 pKa = 11.84AIAAGRR129 pKa = 11.84VALTAGPIGAIRR141 pKa = 11.84ASATRR146 pKa = 11.84SLAIALPRR154 pKa = 11.84LTATRR159 pKa = 11.84GLAVPLPGLVATRR172 pKa = 11.84GLAVPPGPIGAFRR185 pKa = 11.84ASTTRR190 pKa = 11.84SLAVPLPRR198 pKa = 11.84AIPSGRR204 pKa = 11.84VALPTGAA211 pKa = 4.38
MM1 pKa = 7.07TRR3 pKa = 11.84SLHH6 pKa = 5.25FVRR9 pKa = 11.84SARR12 pKa = 11.84PHH14 pKa = 6.65ALLACAGAPSRR25 pKa = 11.84AVAPLGPRR33 pKa = 11.84TARR36 pKa = 11.84RR37 pKa = 11.84VIAPRR42 pKa = 11.84TVVARR47 pKa = 11.84RR48 pKa = 11.84IPVALPGAVAARR60 pKa = 11.84GAVLLPRR67 pKa = 11.84AVASRR72 pKa = 11.84RR73 pKa = 11.84IAVTARR79 pKa = 11.84TIGAVRR85 pKa = 11.84PCPARR90 pKa = 11.84SIAVTLPRR98 pKa = 11.84LIATRR103 pKa = 11.84SLAIPLPRR111 pKa = 11.84PVAARR116 pKa = 11.84RR117 pKa = 11.84AVLLPRR123 pKa = 11.84AIAAGRR129 pKa = 11.84VALTAGPIGAIRR141 pKa = 11.84ASATRR146 pKa = 11.84SLAIALPRR154 pKa = 11.84LTATRR159 pKa = 11.84GLAVPLPGLVATRR172 pKa = 11.84GLAVPPGPIGAFRR185 pKa = 11.84ASTTRR190 pKa = 11.84SLAVPLPRR198 pKa = 11.84AIPSGRR204 pKa = 11.84VALPTGAA211 pKa = 4.38
Molecular weight: 21.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
775821 |
27 |
1968 |
324.7 |
34.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.215 ± 0.084 | 0.457 ± 0.012 |
6.114 ± 0.043 | 6.033 ± 0.04 |
2.922 ± 0.029 | 8.999 ± 0.045 |
1.943 ± 0.022 | 4.643 ± 0.031 |
1.644 ± 0.039 | 10.393 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.729 ± 0.019 | 1.698 ± 0.028 |
5.569 ± 0.041 | 2.559 ± 0.029 |
7.629 ± 0.051 | 5.551 ± 0.03 |
5.678 ± 0.038 | 9.0 ± 0.048 |
1.385 ± 0.022 | 1.838 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
