
Fusarium poae mitovirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.88
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B4ZA48|A0A1B4ZA48_9VIRU RNA-dependent RNA polymerase OS=Fusarium poae mitovirus 2 OX=1848151 PE=4 SV=1
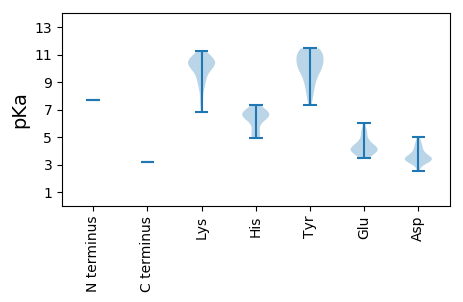
MM1 pKa = 7.68CPLKK5 pKa = 10.95GLTHH9 pKa = 7.33RR10 pKa = 11.84FGGMHH15 pKa = 5.25ITSRR19 pKa = 11.84YY20 pKa = 7.9VFMLITYY27 pKa = 9.0CGRR30 pKa = 11.84NKK32 pKa = 10.61LLIIYY37 pKa = 7.36MKK39 pKa = 8.36NTKK42 pKa = 9.99IIIVLIKK49 pKa = 10.43RR50 pKa = 11.84LILGIFGVDD59 pKa = 3.08HH60 pKa = 6.79TKK62 pKa = 11.05LITEE66 pKa = 4.69FVKK69 pKa = 10.42EE70 pKa = 4.17VQKK73 pKa = 11.14LWINNGFMYY82 pKa = 9.7MISYY86 pKa = 8.11MKK88 pKa = 10.17VVRR91 pKa = 11.84LHH93 pKa = 4.94ITRR96 pKa = 11.84YY97 pKa = 9.17VCGKK101 pKa = 9.03PLYY104 pKa = 10.39SNASNVSLDD113 pKa = 3.28SSGFPTRR120 pKa = 11.84FLFLKK125 pKa = 10.46HH126 pKa = 6.45LLDD129 pKa = 4.99EE130 pKa = 5.27GDD132 pKa = 3.5TGIKK136 pKa = 9.74IVFTLLSLTRR146 pKa = 11.84GLKK149 pKa = 7.51PTKK152 pKa = 10.79AEE154 pKa = 4.12DD155 pKa = 3.33KK156 pKa = 10.54KK157 pKa = 10.94IKK159 pKa = 10.22YY160 pKa = 9.92DD161 pKa = 4.07LGPIIAPHH169 pKa = 6.91KK170 pKa = 7.73GTTMGSVPGWFIKK183 pKa = 10.63EE184 pKa = 3.54FVRR187 pKa = 11.84RR188 pKa = 11.84NNLGRR193 pKa = 11.84KK194 pKa = 8.68LPEE197 pKa = 3.97YY198 pKa = 10.23SVKK201 pKa = 10.75DD202 pKa = 3.39HH203 pKa = 6.28YY204 pKa = 11.44LSTKK208 pKa = 9.95GGPSGKK214 pKa = 8.31STWASQWSHH223 pKa = 6.52LFYY226 pKa = 10.94KK227 pKa = 10.26QDD229 pKa = 4.6LILSLAYY236 pKa = 9.65ILGEE240 pKa = 3.98GFKK243 pKa = 10.55EE244 pKa = 3.94LFYY247 pKa = 11.11TPFLKK252 pKa = 10.67NMHH255 pKa = 6.72LSYY258 pKa = 11.29GKK260 pKa = 10.08DD261 pKa = 2.5RR262 pKa = 11.84WPNGKK267 pKa = 9.96LSIVKK272 pKa = 10.22DD273 pKa = 3.7PEE275 pKa = 3.83GKK277 pKa = 9.77RR278 pKa = 11.84RR279 pKa = 11.84VIAMVDD285 pKa = 3.33YY286 pKa = 10.65HH287 pKa = 6.73SQLALRR293 pKa = 11.84KK294 pKa = 8.81VHH296 pKa = 6.99EE297 pKa = 5.51DD298 pKa = 3.32LLSMLGKK305 pKa = 10.44FSTDD309 pKa = 2.5RR310 pKa = 11.84TFSQNPRR317 pKa = 11.84HH318 pKa = 5.41NWNYY322 pKa = 10.06NNSEE326 pKa = 4.24LFYY329 pKa = 11.28SLDD332 pKa = 3.5LSSATDD338 pKa = 3.73RR339 pKa = 11.84FPVRR343 pKa = 11.84LQARR347 pKa = 11.84LVGEE351 pKa = 4.02IYY353 pKa = 10.47GNHH356 pKa = 6.54RR357 pKa = 11.84FGEE360 pKa = 4.15QWANLLLNRR369 pKa = 11.84DD370 pKa = 3.61YY371 pKa = 10.91MDD373 pKa = 4.76PEE375 pKa = 4.77GNNCRR380 pKa = 11.84YY381 pKa = 10.08AVGQPMGAYY390 pKa = 9.89SSWAAFTLTHH400 pKa = 6.71HH401 pKa = 6.57LTVAWSAYY409 pKa = 8.91KK410 pKa = 10.27ARR412 pKa = 11.84KK413 pKa = 7.42TMGFDD418 pKa = 3.13QYY420 pKa = 11.18IILGDD425 pKa = 4.34DD426 pKa = 3.46IVIKK430 pKa = 10.18DD431 pKa = 3.45NRR433 pKa = 11.84IAEE436 pKa = 4.23IYY438 pKa = 8.68KK439 pKa = 9.16GQMMRR444 pKa = 11.84MGVDD448 pKa = 3.13ISLPKK453 pKa = 9.24THH455 pKa = 6.39VSVDD459 pKa = 3.13TYY461 pKa = 11.34EE462 pKa = 4.03FAKK465 pKa = 10.39RR466 pKa = 11.84WIKK469 pKa = 10.27KK470 pKa = 9.61DD471 pKa = 3.55RR472 pKa = 11.84EE473 pKa = 4.03ITGIPLKK480 pKa = 10.97GILNNINNLKK490 pKa = 10.13IVFTILNDD498 pKa = 3.77YY499 pKa = 10.33LIKK502 pKa = 10.87CPTSVPKK509 pKa = 10.77SSWQIFQSIFLGFQIRR525 pKa = 11.84GGSKK529 pKa = 9.23SRR531 pKa = 11.84KK532 pKa = 6.84TRR534 pKa = 11.84TITKK538 pKa = 9.89KK539 pKa = 10.18YY540 pKa = 10.55LEE542 pKa = 4.23NLRR545 pKa = 11.84DD546 pKa = 3.44FAISVRR552 pKa = 11.84YY553 pKa = 9.78SMNLISPYY561 pKa = 8.38EE562 pKa = 3.79LRR564 pKa = 11.84AYY566 pKa = 9.27FASKK570 pKa = 10.61VKK572 pKa = 10.53LIKK575 pKa = 9.36EE576 pKa = 4.18TSDD579 pKa = 3.46YY580 pKa = 11.3QSIPSEE586 pKa = 4.13KK587 pKa = 10.6LILQYY592 pKa = 10.18MEE594 pKa = 6.05GILTNGLAKK603 pKa = 10.11ISKK606 pKa = 8.73DD607 pKa = 3.42TIIQINKK614 pKa = 9.09QLDD617 pKa = 3.61SFDD620 pKa = 4.37RR621 pKa = 11.84LAKK624 pKa = 9.41EE625 pKa = 4.5DD626 pKa = 3.88KK627 pKa = 10.5RR628 pKa = 11.84SLVYY632 pKa = 10.68SGVLHH637 pKa = 6.66GLMNRR642 pKa = 11.84LEE644 pKa = 4.45RR645 pKa = 11.84LQEE648 pKa = 3.65LCQRR652 pKa = 11.84MKK654 pKa = 11.25DD655 pKa = 3.39EE656 pKa = 4.63GSTVVEE662 pKa = 5.45FINHH666 pKa = 4.99FTAPSVDD673 pKa = 3.28SLSRR677 pKa = 11.84KK678 pKa = 9.93DD679 pKa = 3.25RR680 pKa = 11.84DD681 pKa = 3.28INIRR685 pKa = 11.84MAFLDD690 pKa = 4.07SLWKK694 pKa = 10.48KK695 pKa = 10.37SLQKK699 pKa = 10.59HH700 pKa = 5.85FSDD703 pKa = 4.24QRR705 pKa = 11.84FPDD708 pKa = 3.13SYY710 pKa = 11.23YY711 pKa = 11.01RR712 pKa = 11.84NLEE715 pKa = 3.59TRR717 pKa = 11.84TLYY720 pKa = 11.08GSLDD724 pKa = 3.51ALDD727 pKa = 4.56IIDD730 pKa = 4.04YY731 pKa = 10.13EE732 pKa = 4.66GEE734 pKa = 3.99IEE736 pKa = 5.22VRR738 pKa = 11.84PIWSRR743 pKa = 11.84EE744 pKa = 3.45LSQFNQRR751 pKa = 11.84SINSLQMFISEE762 pKa = 4.22NSS764 pKa = 3.17
MM1 pKa = 7.68CPLKK5 pKa = 10.95GLTHH9 pKa = 7.33RR10 pKa = 11.84FGGMHH15 pKa = 5.25ITSRR19 pKa = 11.84YY20 pKa = 7.9VFMLITYY27 pKa = 9.0CGRR30 pKa = 11.84NKK32 pKa = 10.61LLIIYY37 pKa = 7.36MKK39 pKa = 8.36NTKK42 pKa = 9.99IIIVLIKK49 pKa = 10.43RR50 pKa = 11.84LILGIFGVDD59 pKa = 3.08HH60 pKa = 6.79TKK62 pKa = 11.05LITEE66 pKa = 4.69FVKK69 pKa = 10.42EE70 pKa = 4.17VQKK73 pKa = 11.14LWINNGFMYY82 pKa = 9.7MISYY86 pKa = 8.11MKK88 pKa = 10.17VVRR91 pKa = 11.84LHH93 pKa = 4.94ITRR96 pKa = 11.84YY97 pKa = 9.17VCGKK101 pKa = 9.03PLYY104 pKa = 10.39SNASNVSLDD113 pKa = 3.28SSGFPTRR120 pKa = 11.84FLFLKK125 pKa = 10.46HH126 pKa = 6.45LLDD129 pKa = 4.99EE130 pKa = 5.27GDD132 pKa = 3.5TGIKK136 pKa = 9.74IVFTLLSLTRR146 pKa = 11.84GLKK149 pKa = 7.51PTKK152 pKa = 10.79AEE154 pKa = 4.12DD155 pKa = 3.33KK156 pKa = 10.54KK157 pKa = 10.94IKK159 pKa = 10.22YY160 pKa = 9.92DD161 pKa = 4.07LGPIIAPHH169 pKa = 6.91KK170 pKa = 7.73GTTMGSVPGWFIKK183 pKa = 10.63EE184 pKa = 3.54FVRR187 pKa = 11.84RR188 pKa = 11.84NNLGRR193 pKa = 11.84KK194 pKa = 8.68LPEE197 pKa = 3.97YY198 pKa = 10.23SVKK201 pKa = 10.75DD202 pKa = 3.39HH203 pKa = 6.28YY204 pKa = 11.44LSTKK208 pKa = 9.95GGPSGKK214 pKa = 8.31STWASQWSHH223 pKa = 6.52LFYY226 pKa = 10.94KK227 pKa = 10.26QDD229 pKa = 4.6LILSLAYY236 pKa = 9.65ILGEE240 pKa = 3.98GFKK243 pKa = 10.55EE244 pKa = 3.94LFYY247 pKa = 11.11TPFLKK252 pKa = 10.67NMHH255 pKa = 6.72LSYY258 pKa = 11.29GKK260 pKa = 10.08DD261 pKa = 2.5RR262 pKa = 11.84WPNGKK267 pKa = 9.96LSIVKK272 pKa = 10.22DD273 pKa = 3.7PEE275 pKa = 3.83GKK277 pKa = 9.77RR278 pKa = 11.84RR279 pKa = 11.84VIAMVDD285 pKa = 3.33YY286 pKa = 10.65HH287 pKa = 6.73SQLALRR293 pKa = 11.84KK294 pKa = 8.81VHH296 pKa = 6.99EE297 pKa = 5.51DD298 pKa = 3.32LLSMLGKK305 pKa = 10.44FSTDD309 pKa = 2.5RR310 pKa = 11.84TFSQNPRR317 pKa = 11.84HH318 pKa = 5.41NWNYY322 pKa = 10.06NNSEE326 pKa = 4.24LFYY329 pKa = 11.28SLDD332 pKa = 3.5LSSATDD338 pKa = 3.73RR339 pKa = 11.84FPVRR343 pKa = 11.84LQARR347 pKa = 11.84LVGEE351 pKa = 4.02IYY353 pKa = 10.47GNHH356 pKa = 6.54RR357 pKa = 11.84FGEE360 pKa = 4.15QWANLLLNRR369 pKa = 11.84DD370 pKa = 3.61YY371 pKa = 10.91MDD373 pKa = 4.76PEE375 pKa = 4.77GNNCRR380 pKa = 11.84YY381 pKa = 10.08AVGQPMGAYY390 pKa = 9.89SSWAAFTLTHH400 pKa = 6.71HH401 pKa = 6.57LTVAWSAYY409 pKa = 8.91KK410 pKa = 10.27ARR412 pKa = 11.84KK413 pKa = 7.42TMGFDD418 pKa = 3.13QYY420 pKa = 11.18IILGDD425 pKa = 4.34DD426 pKa = 3.46IVIKK430 pKa = 10.18DD431 pKa = 3.45NRR433 pKa = 11.84IAEE436 pKa = 4.23IYY438 pKa = 8.68KK439 pKa = 9.16GQMMRR444 pKa = 11.84MGVDD448 pKa = 3.13ISLPKK453 pKa = 9.24THH455 pKa = 6.39VSVDD459 pKa = 3.13TYY461 pKa = 11.34EE462 pKa = 4.03FAKK465 pKa = 10.39RR466 pKa = 11.84WIKK469 pKa = 10.27KK470 pKa = 9.61DD471 pKa = 3.55RR472 pKa = 11.84EE473 pKa = 4.03ITGIPLKK480 pKa = 10.97GILNNINNLKK490 pKa = 10.13IVFTILNDD498 pKa = 3.77YY499 pKa = 10.33LIKK502 pKa = 10.87CPTSVPKK509 pKa = 10.77SSWQIFQSIFLGFQIRR525 pKa = 11.84GGSKK529 pKa = 9.23SRR531 pKa = 11.84KK532 pKa = 6.84TRR534 pKa = 11.84TITKK538 pKa = 9.89KK539 pKa = 10.18YY540 pKa = 10.55LEE542 pKa = 4.23NLRR545 pKa = 11.84DD546 pKa = 3.44FAISVRR552 pKa = 11.84YY553 pKa = 9.78SMNLISPYY561 pKa = 8.38EE562 pKa = 3.79LRR564 pKa = 11.84AYY566 pKa = 9.27FASKK570 pKa = 10.61VKK572 pKa = 10.53LIKK575 pKa = 9.36EE576 pKa = 4.18TSDD579 pKa = 3.46YY580 pKa = 11.3QSIPSEE586 pKa = 4.13KK587 pKa = 10.6LILQYY592 pKa = 10.18MEE594 pKa = 6.05GILTNGLAKK603 pKa = 10.11ISKK606 pKa = 8.73DD607 pKa = 3.42TIIQINKK614 pKa = 9.09QLDD617 pKa = 3.61SFDD620 pKa = 4.37RR621 pKa = 11.84LAKK624 pKa = 9.41EE625 pKa = 4.5DD626 pKa = 3.88KK627 pKa = 10.5RR628 pKa = 11.84SLVYY632 pKa = 10.68SGVLHH637 pKa = 6.66GLMNRR642 pKa = 11.84LEE644 pKa = 4.45RR645 pKa = 11.84LQEE648 pKa = 3.65LCQRR652 pKa = 11.84MKK654 pKa = 11.25DD655 pKa = 3.39EE656 pKa = 4.63GSTVVEE662 pKa = 5.45FINHH666 pKa = 4.99FTAPSVDD673 pKa = 3.28SLSRR677 pKa = 11.84KK678 pKa = 9.93DD679 pKa = 3.25RR680 pKa = 11.84DD681 pKa = 3.28INIRR685 pKa = 11.84MAFLDD690 pKa = 4.07SLWKK694 pKa = 10.48KK695 pKa = 10.37SLQKK699 pKa = 10.59HH700 pKa = 5.85FSDD703 pKa = 4.24QRR705 pKa = 11.84FPDD708 pKa = 3.13SYY710 pKa = 11.23YY711 pKa = 11.01RR712 pKa = 11.84NLEE715 pKa = 3.59TRR717 pKa = 11.84TLYY720 pKa = 11.08GSLDD724 pKa = 3.51ALDD727 pKa = 4.56IIDD730 pKa = 4.04YY731 pKa = 10.13EE732 pKa = 4.66GEE734 pKa = 3.99IEE736 pKa = 5.22VRR738 pKa = 11.84PIWSRR743 pKa = 11.84EE744 pKa = 3.45LSQFNQRR751 pKa = 11.84SINSLQMFISEE762 pKa = 4.22NSS764 pKa = 3.17
Molecular weight: 88.5 kDa
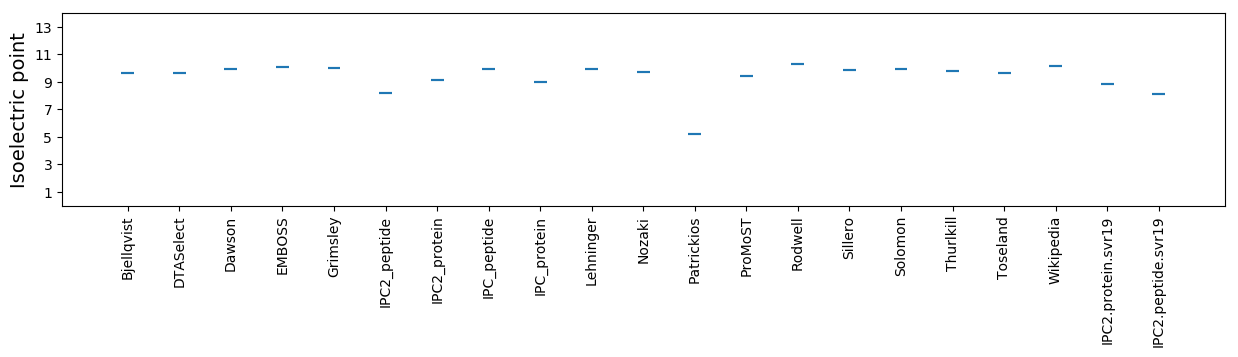
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B4ZA48|A0A1B4ZA48_9VIRU RNA-dependent RNA polymerase OS=Fusarium poae mitovirus 2 OX=1848151 PE=4 SV=1
MM1 pKa = 7.68CPLKK5 pKa = 10.95GLTHH9 pKa = 7.33RR10 pKa = 11.84FGGMHH15 pKa = 5.25ITSRR19 pKa = 11.84YY20 pKa = 7.9VFMLITYY27 pKa = 9.0CGRR30 pKa = 11.84NKK32 pKa = 10.61LLIIYY37 pKa = 7.36MKK39 pKa = 8.36NTKK42 pKa = 9.99IIIVLIKK49 pKa = 10.43RR50 pKa = 11.84LILGIFGVDD59 pKa = 3.08HH60 pKa = 6.79TKK62 pKa = 11.05LITEE66 pKa = 4.69FVKK69 pKa = 10.42EE70 pKa = 4.17VQKK73 pKa = 11.14LWINNGFMYY82 pKa = 9.7MISYY86 pKa = 8.11MKK88 pKa = 10.17VVRR91 pKa = 11.84LHH93 pKa = 4.94ITRR96 pKa = 11.84YY97 pKa = 9.17VCGKK101 pKa = 9.03PLYY104 pKa = 10.39SNASNVSLDD113 pKa = 3.28SSGFPTRR120 pKa = 11.84FLFLKK125 pKa = 10.46HH126 pKa = 6.45LLDD129 pKa = 4.99EE130 pKa = 5.27GDD132 pKa = 3.5TGIKK136 pKa = 9.74IVFTLLSLTRR146 pKa = 11.84GLKK149 pKa = 7.51PTKK152 pKa = 10.79AEE154 pKa = 4.12DD155 pKa = 3.33KK156 pKa = 10.54KK157 pKa = 10.94IKK159 pKa = 10.22YY160 pKa = 9.92DD161 pKa = 4.07LGPIIAPHH169 pKa = 6.91KK170 pKa = 7.73GTTMGSVPGWFIKK183 pKa = 10.63EE184 pKa = 3.54FVRR187 pKa = 11.84RR188 pKa = 11.84NNLGRR193 pKa = 11.84KK194 pKa = 8.68LPEE197 pKa = 3.97YY198 pKa = 10.23SVKK201 pKa = 10.75DD202 pKa = 3.39HH203 pKa = 6.28YY204 pKa = 11.44LSTKK208 pKa = 9.95GGPSGKK214 pKa = 8.31STWASQWSHH223 pKa = 6.52LFYY226 pKa = 10.94KK227 pKa = 10.26QDD229 pKa = 4.6LILSLAYY236 pKa = 9.65ILGEE240 pKa = 3.98GFKK243 pKa = 10.55EE244 pKa = 3.94LFYY247 pKa = 11.11TPFLKK252 pKa = 10.67NMHH255 pKa = 6.72LSYY258 pKa = 11.29GKK260 pKa = 10.08DD261 pKa = 2.5RR262 pKa = 11.84WPNGKK267 pKa = 9.96LSIVKK272 pKa = 10.22DD273 pKa = 3.7PEE275 pKa = 3.83GKK277 pKa = 9.77RR278 pKa = 11.84RR279 pKa = 11.84VIAMVDD285 pKa = 3.33YY286 pKa = 10.65HH287 pKa = 6.73SQLALRR293 pKa = 11.84KK294 pKa = 8.81VHH296 pKa = 6.99EE297 pKa = 5.51DD298 pKa = 3.32LLSMLGKK305 pKa = 10.44FSTDD309 pKa = 2.5RR310 pKa = 11.84TFSQNPRR317 pKa = 11.84HH318 pKa = 5.41NWNYY322 pKa = 10.06NNSEE326 pKa = 4.24LFYY329 pKa = 11.28SLDD332 pKa = 3.5LSSATDD338 pKa = 3.73RR339 pKa = 11.84FPVRR343 pKa = 11.84LQARR347 pKa = 11.84LVGEE351 pKa = 4.02IYY353 pKa = 10.47GNHH356 pKa = 6.54RR357 pKa = 11.84FGEE360 pKa = 4.15QWANLLLNRR369 pKa = 11.84DD370 pKa = 3.61YY371 pKa = 10.91MDD373 pKa = 4.76PEE375 pKa = 4.77GNNCRR380 pKa = 11.84YY381 pKa = 10.08AVGQPMGAYY390 pKa = 9.89SSWAAFTLTHH400 pKa = 6.71HH401 pKa = 6.57LTVAWSAYY409 pKa = 8.91KK410 pKa = 10.27ARR412 pKa = 11.84KK413 pKa = 7.42TMGFDD418 pKa = 3.13QYY420 pKa = 11.18IILGDD425 pKa = 4.34DD426 pKa = 3.46IVIKK430 pKa = 10.18DD431 pKa = 3.45NRR433 pKa = 11.84IAEE436 pKa = 4.23IYY438 pKa = 8.68KK439 pKa = 9.16GQMMRR444 pKa = 11.84MGVDD448 pKa = 3.13ISLPKK453 pKa = 9.24THH455 pKa = 6.39VSVDD459 pKa = 3.13TYY461 pKa = 11.34EE462 pKa = 4.03FAKK465 pKa = 10.39RR466 pKa = 11.84WIKK469 pKa = 10.27KK470 pKa = 9.61DD471 pKa = 3.55RR472 pKa = 11.84EE473 pKa = 4.03ITGIPLKK480 pKa = 10.97GILNNINNLKK490 pKa = 10.13IVFTILNDD498 pKa = 3.77YY499 pKa = 10.33LIKK502 pKa = 10.87CPTSVPKK509 pKa = 10.77SSWQIFQSIFLGFQIRR525 pKa = 11.84GGSKK529 pKa = 9.23SRR531 pKa = 11.84KK532 pKa = 6.84TRR534 pKa = 11.84TITKK538 pKa = 9.89KK539 pKa = 10.18YY540 pKa = 10.55LEE542 pKa = 4.23NLRR545 pKa = 11.84DD546 pKa = 3.44FAISVRR552 pKa = 11.84YY553 pKa = 9.78SMNLISPYY561 pKa = 8.38EE562 pKa = 3.79LRR564 pKa = 11.84AYY566 pKa = 9.27FASKK570 pKa = 10.61VKK572 pKa = 10.53LIKK575 pKa = 9.36EE576 pKa = 4.18TSDD579 pKa = 3.46YY580 pKa = 11.3QSIPSEE586 pKa = 4.13KK587 pKa = 10.6LILQYY592 pKa = 10.18MEE594 pKa = 6.05GILTNGLAKK603 pKa = 10.11ISKK606 pKa = 8.73DD607 pKa = 3.42TIIQINKK614 pKa = 9.09QLDD617 pKa = 3.61SFDD620 pKa = 4.37RR621 pKa = 11.84LAKK624 pKa = 9.41EE625 pKa = 4.5DD626 pKa = 3.88KK627 pKa = 10.5RR628 pKa = 11.84SLVYY632 pKa = 10.68SGVLHH637 pKa = 6.66GLMNRR642 pKa = 11.84LEE644 pKa = 4.45RR645 pKa = 11.84LQEE648 pKa = 3.65LCQRR652 pKa = 11.84MKK654 pKa = 11.25DD655 pKa = 3.39EE656 pKa = 4.63GSTVVEE662 pKa = 5.45FINHH666 pKa = 4.99FTAPSVDD673 pKa = 3.28SLSRR677 pKa = 11.84KK678 pKa = 9.93DD679 pKa = 3.25RR680 pKa = 11.84DD681 pKa = 3.28INIRR685 pKa = 11.84MAFLDD690 pKa = 4.07SLWKK694 pKa = 10.48KK695 pKa = 10.37SLQKK699 pKa = 10.59HH700 pKa = 5.85FSDD703 pKa = 4.24QRR705 pKa = 11.84FPDD708 pKa = 3.13SYY710 pKa = 11.23YY711 pKa = 11.01RR712 pKa = 11.84NLEE715 pKa = 3.59TRR717 pKa = 11.84TLYY720 pKa = 11.08GSLDD724 pKa = 3.51ALDD727 pKa = 4.56IIDD730 pKa = 4.04YY731 pKa = 10.13EE732 pKa = 4.66GEE734 pKa = 3.99IEE736 pKa = 5.22VRR738 pKa = 11.84PIWSRR743 pKa = 11.84EE744 pKa = 3.45LSQFNQRR751 pKa = 11.84SINSLQMFISEE762 pKa = 4.22NSS764 pKa = 3.17
MM1 pKa = 7.68CPLKK5 pKa = 10.95GLTHH9 pKa = 7.33RR10 pKa = 11.84FGGMHH15 pKa = 5.25ITSRR19 pKa = 11.84YY20 pKa = 7.9VFMLITYY27 pKa = 9.0CGRR30 pKa = 11.84NKK32 pKa = 10.61LLIIYY37 pKa = 7.36MKK39 pKa = 8.36NTKK42 pKa = 9.99IIIVLIKK49 pKa = 10.43RR50 pKa = 11.84LILGIFGVDD59 pKa = 3.08HH60 pKa = 6.79TKK62 pKa = 11.05LITEE66 pKa = 4.69FVKK69 pKa = 10.42EE70 pKa = 4.17VQKK73 pKa = 11.14LWINNGFMYY82 pKa = 9.7MISYY86 pKa = 8.11MKK88 pKa = 10.17VVRR91 pKa = 11.84LHH93 pKa = 4.94ITRR96 pKa = 11.84YY97 pKa = 9.17VCGKK101 pKa = 9.03PLYY104 pKa = 10.39SNASNVSLDD113 pKa = 3.28SSGFPTRR120 pKa = 11.84FLFLKK125 pKa = 10.46HH126 pKa = 6.45LLDD129 pKa = 4.99EE130 pKa = 5.27GDD132 pKa = 3.5TGIKK136 pKa = 9.74IVFTLLSLTRR146 pKa = 11.84GLKK149 pKa = 7.51PTKK152 pKa = 10.79AEE154 pKa = 4.12DD155 pKa = 3.33KK156 pKa = 10.54KK157 pKa = 10.94IKK159 pKa = 10.22YY160 pKa = 9.92DD161 pKa = 4.07LGPIIAPHH169 pKa = 6.91KK170 pKa = 7.73GTTMGSVPGWFIKK183 pKa = 10.63EE184 pKa = 3.54FVRR187 pKa = 11.84RR188 pKa = 11.84NNLGRR193 pKa = 11.84KK194 pKa = 8.68LPEE197 pKa = 3.97YY198 pKa = 10.23SVKK201 pKa = 10.75DD202 pKa = 3.39HH203 pKa = 6.28YY204 pKa = 11.44LSTKK208 pKa = 9.95GGPSGKK214 pKa = 8.31STWASQWSHH223 pKa = 6.52LFYY226 pKa = 10.94KK227 pKa = 10.26QDD229 pKa = 4.6LILSLAYY236 pKa = 9.65ILGEE240 pKa = 3.98GFKK243 pKa = 10.55EE244 pKa = 3.94LFYY247 pKa = 11.11TPFLKK252 pKa = 10.67NMHH255 pKa = 6.72LSYY258 pKa = 11.29GKK260 pKa = 10.08DD261 pKa = 2.5RR262 pKa = 11.84WPNGKK267 pKa = 9.96LSIVKK272 pKa = 10.22DD273 pKa = 3.7PEE275 pKa = 3.83GKK277 pKa = 9.77RR278 pKa = 11.84RR279 pKa = 11.84VIAMVDD285 pKa = 3.33YY286 pKa = 10.65HH287 pKa = 6.73SQLALRR293 pKa = 11.84KK294 pKa = 8.81VHH296 pKa = 6.99EE297 pKa = 5.51DD298 pKa = 3.32LLSMLGKK305 pKa = 10.44FSTDD309 pKa = 2.5RR310 pKa = 11.84TFSQNPRR317 pKa = 11.84HH318 pKa = 5.41NWNYY322 pKa = 10.06NNSEE326 pKa = 4.24LFYY329 pKa = 11.28SLDD332 pKa = 3.5LSSATDD338 pKa = 3.73RR339 pKa = 11.84FPVRR343 pKa = 11.84LQARR347 pKa = 11.84LVGEE351 pKa = 4.02IYY353 pKa = 10.47GNHH356 pKa = 6.54RR357 pKa = 11.84FGEE360 pKa = 4.15QWANLLLNRR369 pKa = 11.84DD370 pKa = 3.61YY371 pKa = 10.91MDD373 pKa = 4.76PEE375 pKa = 4.77GNNCRR380 pKa = 11.84YY381 pKa = 10.08AVGQPMGAYY390 pKa = 9.89SSWAAFTLTHH400 pKa = 6.71HH401 pKa = 6.57LTVAWSAYY409 pKa = 8.91KK410 pKa = 10.27ARR412 pKa = 11.84KK413 pKa = 7.42TMGFDD418 pKa = 3.13QYY420 pKa = 11.18IILGDD425 pKa = 4.34DD426 pKa = 3.46IVIKK430 pKa = 10.18DD431 pKa = 3.45NRR433 pKa = 11.84IAEE436 pKa = 4.23IYY438 pKa = 8.68KK439 pKa = 9.16GQMMRR444 pKa = 11.84MGVDD448 pKa = 3.13ISLPKK453 pKa = 9.24THH455 pKa = 6.39VSVDD459 pKa = 3.13TYY461 pKa = 11.34EE462 pKa = 4.03FAKK465 pKa = 10.39RR466 pKa = 11.84WIKK469 pKa = 10.27KK470 pKa = 9.61DD471 pKa = 3.55RR472 pKa = 11.84EE473 pKa = 4.03ITGIPLKK480 pKa = 10.97GILNNINNLKK490 pKa = 10.13IVFTILNDD498 pKa = 3.77YY499 pKa = 10.33LIKK502 pKa = 10.87CPTSVPKK509 pKa = 10.77SSWQIFQSIFLGFQIRR525 pKa = 11.84GGSKK529 pKa = 9.23SRR531 pKa = 11.84KK532 pKa = 6.84TRR534 pKa = 11.84TITKK538 pKa = 9.89KK539 pKa = 10.18YY540 pKa = 10.55LEE542 pKa = 4.23NLRR545 pKa = 11.84DD546 pKa = 3.44FAISVRR552 pKa = 11.84YY553 pKa = 9.78SMNLISPYY561 pKa = 8.38EE562 pKa = 3.79LRR564 pKa = 11.84AYY566 pKa = 9.27FASKK570 pKa = 10.61VKK572 pKa = 10.53LIKK575 pKa = 9.36EE576 pKa = 4.18TSDD579 pKa = 3.46YY580 pKa = 11.3QSIPSEE586 pKa = 4.13KK587 pKa = 10.6LILQYY592 pKa = 10.18MEE594 pKa = 6.05GILTNGLAKK603 pKa = 10.11ISKK606 pKa = 8.73DD607 pKa = 3.42TIIQINKK614 pKa = 9.09QLDD617 pKa = 3.61SFDD620 pKa = 4.37RR621 pKa = 11.84LAKK624 pKa = 9.41EE625 pKa = 4.5DD626 pKa = 3.88KK627 pKa = 10.5RR628 pKa = 11.84SLVYY632 pKa = 10.68SGVLHH637 pKa = 6.66GLMNRR642 pKa = 11.84LEE644 pKa = 4.45RR645 pKa = 11.84LQEE648 pKa = 3.65LCQRR652 pKa = 11.84MKK654 pKa = 11.25DD655 pKa = 3.39EE656 pKa = 4.63GSTVVEE662 pKa = 5.45FINHH666 pKa = 4.99FTAPSVDD673 pKa = 3.28SLSRR677 pKa = 11.84KK678 pKa = 9.93DD679 pKa = 3.25RR680 pKa = 11.84DD681 pKa = 3.28INIRR685 pKa = 11.84MAFLDD690 pKa = 4.07SLWKK694 pKa = 10.48KK695 pKa = 10.37SLQKK699 pKa = 10.59HH700 pKa = 5.85FSDD703 pKa = 4.24QRR705 pKa = 11.84FPDD708 pKa = 3.13SYY710 pKa = 11.23YY711 pKa = 11.01RR712 pKa = 11.84NLEE715 pKa = 3.59TRR717 pKa = 11.84TLYY720 pKa = 11.08GSLDD724 pKa = 3.51ALDD727 pKa = 4.56IIDD730 pKa = 4.04YY731 pKa = 10.13EE732 pKa = 4.66GEE734 pKa = 3.99IEE736 pKa = 5.22VRR738 pKa = 11.84PIWSRR743 pKa = 11.84EE744 pKa = 3.45LSQFNQRR751 pKa = 11.84SINSLQMFISEE762 pKa = 4.22NSS764 pKa = 3.17
Molecular weight: 88.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
764 |
764 |
764 |
764.0 |
88.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.534 ± 0.0 | 0.785 ± 0.0 |
5.366 ± 0.0 | 4.319 ± 0.0 |
4.843 ± 0.0 | 6.283 ± 0.0 |
2.487 ± 0.0 | 8.246 ± 0.0 |
8.115 ± 0.0 | 10.995 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.01 ± 0.0 | 4.843 ± 0.0 |
3.272 ± 0.0 | 3.141 ± 0.0 |
6.283 ± 0.0 | 8.246 ± 0.0 |
5.105 ± 0.0 | 4.581 ± 0.0 |
1.702 ± 0.0 | 4.843 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
