
Cabbage cytorhabdovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Cytorhabdovirus; Cabbage cytorhabdovirus
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
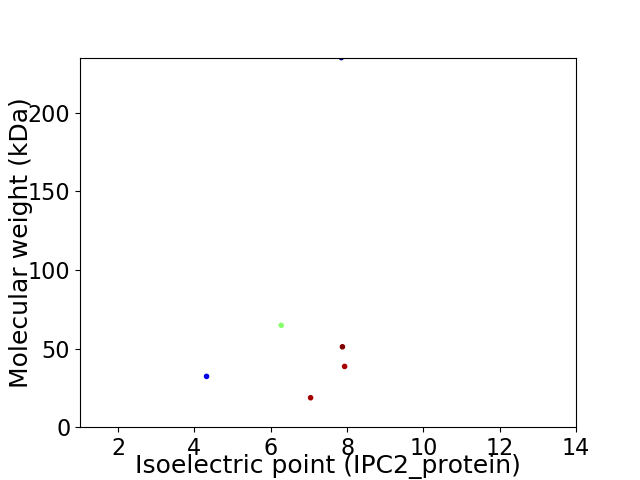
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
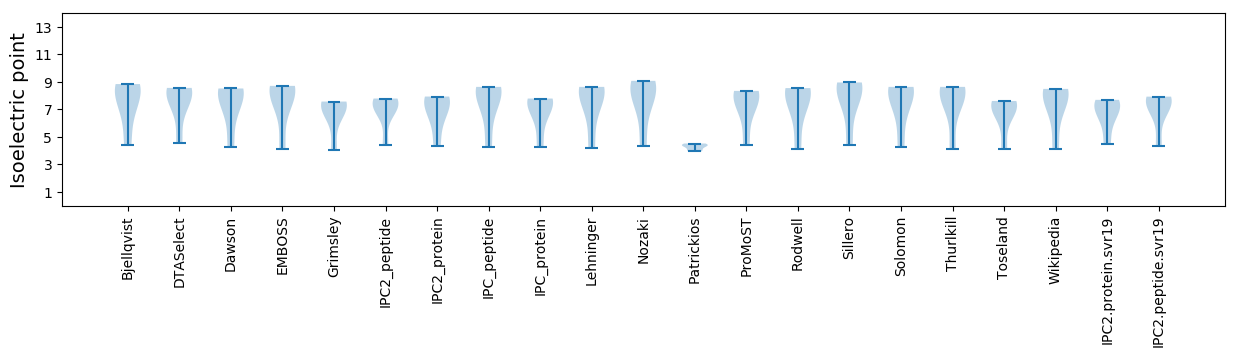
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2PYM8|A0A2D2PYM8_9RHAB Matrix protein OS=Cabbage cytorhabdovirus 1 OX=2051550 PE=4 SV=1
MM1 pKa = 8.2DD2 pKa = 5.15PQQSAIDD9 pKa = 3.93FSEE12 pKa = 4.04IDD14 pKa = 3.77ANLHH18 pKa = 5.41RR19 pKa = 11.84SPYY22 pKa = 9.77VVPALDD28 pKa = 4.57DD29 pKa = 4.8DD30 pKa = 4.14MDD32 pKa = 5.31GGEE35 pKa = 4.44IVQDD39 pKa = 3.8DD40 pKa = 4.33LSVPRR45 pKa = 11.84STDD48 pKa = 3.03DD49 pKa = 3.82VAPPTQKK56 pKa = 10.69SSLVSSEE63 pKa = 4.07VVEE66 pKa = 4.24SLLYY70 pKa = 9.76EE71 pKa = 4.06ASVLHH76 pKa = 6.67GIAVTPHH83 pKa = 6.12MSTTAIALAHH93 pKa = 6.3NIGLEE98 pKa = 4.01PHH100 pKa = 6.04SLDD103 pKa = 3.04WFLAGISYY111 pKa = 10.83ANNSMIVEE119 pKa = 4.27KK120 pKa = 10.49LSSIIKK126 pKa = 10.22DD127 pKa = 3.45MQIEE131 pKa = 4.31TRR133 pKa = 11.84NLQTATSSVAAVSNEE148 pKa = 3.48FLGKK152 pKa = 9.22MSRR155 pKa = 11.84NKK157 pKa = 10.45RR158 pKa = 11.84EE159 pKa = 4.52IIDD162 pKa = 3.57EE163 pKa = 4.15MEE165 pKa = 4.04KK166 pKa = 9.78TRR168 pKa = 11.84EE169 pKa = 4.04SVLNAVASIQSAEE182 pKa = 4.19LGSHH186 pKa = 6.62IGDD189 pKa = 3.74DD190 pKa = 4.26FVNLTVNPEE199 pKa = 4.08GTSEE203 pKa = 4.28RR204 pKa = 11.84STVPPPAPVDD214 pKa = 3.33AGLLSKK220 pKa = 10.77VLSNPVTLKK229 pKa = 11.03SPDD232 pKa = 3.5EE233 pKa = 3.54ILYY236 pKa = 9.68ARR238 pKa = 11.84KK239 pKa = 9.93YY240 pKa = 10.84KK241 pKa = 10.77LLLDD245 pKa = 4.55LGIEE249 pKa = 3.97IPKK252 pKa = 10.69ANLSPDD258 pKa = 2.95IMNTLIPDD266 pKa = 3.82WQLEE270 pKa = 4.08AAEE273 pKa = 5.08AGLDD277 pKa = 3.59SKK279 pKa = 10.12TKK281 pKa = 10.09MEE283 pKa = 4.49LTDD286 pKa = 3.78EE287 pKa = 4.63LLDD290 pKa = 4.34IVITLNLII298 pKa = 3.82
MM1 pKa = 8.2DD2 pKa = 5.15PQQSAIDD9 pKa = 3.93FSEE12 pKa = 4.04IDD14 pKa = 3.77ANLHH18 pKa = 5.41RR19 pKa = 11.84SPYY22 pKa = 9.77VVPALDD28 pKa = 4.57DD29 pKa = 4.8DD30 pKa = 4.14MDD32 pKa = 5.31GGEE35 pKa = 4.44IVQDD39 pKa = 3.8DD40 pKa = 4.33LSVPRR45 pKa = 11.84STDD48 pKa = 3.03DD49 pKa = 3.82VAPPTQKK56 pKa = 10.69SSLVSSEE63 pKa = 4.07VVEE66 pKa = 4.24SLLYY70 pKa = 9.76EE71 pKa = 4.06ASVLHH76 pKa = 6.67GIAVTPHH83 pKa = 6.12MSTTAIALAHH93 pKa = 6.3NIGLEE98 pKa = 4.01PHH100 pKa = 6.04SLDD103 pKa = 3.04WFLAGISYY111 pKa = 10.83ANNSMIVEE119 pKa = 4.27KK120 pKa = 10.49LSSIIKK126 pKa = 10.22DD127 pKa = 3.45MQIEE131 pKa = 4.31TRR133 pKa = 11.84NLQTATSSVAAVSNEE148 pKa = 3.48FLGKK152 pKa = 9.22MSRR155 pKa = 11.84NKK157 pKa = 10.45RR158 pKa = 11.84EE159 pKa = 4.52IIDD162 pKa = 3.57EE163 pKa = 4.15MEE165 pKa = 4.04KK166 pKa = 9.78TRR168 pKa = 11.84EE169 pKa = 4.04SVLNAVASIQSAEE182 pKa = 4.19LGSHH186 pKa = 6.62IGDD189 pKa = 3.74DD190 pKa = 4.26FVNLTVNPEE199 pKa = 4.08GTSEE203 pKa = 4.28RR204 pKa = 11.84STVPPPAPVDD214 pKa = 3.33AGLLSKK220 pKa = 10.77VLSNPVTLKK229 pKa = 11.03SPDD232 pKa = 3.5EE233 pKa = 3.54ILYY236 pKa = 9.68ARR238 pKa = 11.84KK239 pKa = 9.93YY240 pKa = 10.84KK241 pKa = 10.77LLLDD245 pKa = 4.55LGIEE249 pKa = 3.97IPKK252 pKa = 10.69ANLSPDD258 pKa = 2.95IMNTLIPDD266 pKa = 3.82WQLEE270 pKa = 4.08AAEE273 pKa = 5.08AGLDD277 pKa = 3.59SKK279 pKa = 10.12TKK281 pKa = 10.09MEE283 pKa = 4.49LTDD286 pKa = 3.78EE287 pKa = 4.63LLDD290 pKa = 4.34IVITLNLII298 pKa = 3.82
Molecular weight: 32.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2PYL4|A0A2D2PYL4_9RHAB Glycoprotein OS=Cabbage cytorhabdovirus 1 OX=2051550 PE=4 SV=1
MM1 pKa = 7.58CLSCLKK7 pKa = 10.28YY8 pKa = 10.72VWNLCLTMCDD18 pKa = 3.07LRR20 pKa = 11.84KK21 pKa = 9.62LVRR24 pKa = 11.84VTMINVNDD32 pKa = 3.49VRR34 pKa = 11.84KK35 pKa = 9.98QIIKK39 pKa = 10.51SGSLTSAVGTGYY51 pKa = 11.1VYY53 pKa = 10.84DD54 pKa = 4.05GSYY57 pKa = 11.05NKK59 pKa = 10.03YY60 pKa = 9.56ARR62 pKa = 11.84KK63 pKa = 9.7RR64 pKa = 11.84EE65 pKa = 3.97LNLRR69 pKa = 11.84VSSSGDD75 pKa = 3.14KK76 pKa = 10.56NIMMRR81 pKa = 11.84HH82 pKa = 4.69VPIFDD87 pKa = 5.11DD88 pKa = 3.67EE89 pKa = 4.4DD90 pKa = 3.6LRR92 pKa = 11.84ALRR95 pKa = 11.84SEE97 pKa = 4.29ANSNKK102 pKa = 9.82YY103 pKa = 9.21IHH105 pKa = 6.85IGCLTISIEE114 pKa = 4.05PLMHH118 pKa = 6.58KK119 pKa = 10.08RR120 pKa = 11.84YY121 pKa = 9.07MDD123 pKa = 4.17LYY125 pKa = 8.05GDD127 pKa = 3.9KK128 pKa = 10.17MRR130 pKa = 11.84GICAVIDD137 pKa = 3.52TTFADD142 pKa = 3.95PAEE145 pKa = 4.84SIISAHH151 pKa = 6.8RR152 pKa = 11.84YY153 pKa = 7.26EE154 pKa = 4.35LSKK157 pKa = 11.12GRR159 pKa = 11.84ADD161 pKa = 3.67FVSMPNHH168 pKa = 6.43CLSLLDD174 pKa = 4.42PNLKK178 pKa = 9.67QRR180 pKa = 11.84LSVMISLDD188 pKa = 3.9GINVKK193 pKa = 10.21KK194 pKa = 10.34GNEE197 pKa = 3.97MFNVCIGYY205 pKa = 9.47IVTGVNTLNPTKK217 pKa = 10.15AVSMSNIPITGTSEE231 pKa = 4.26CEE233 pKa = 3.9PGDD236 pKa = 3.69LSEE239 pKa = 6.36DD240 pKa = 3.29MLEE243 pKa = 4.66GIKK246 pKa = 10.69GSNGNLLISHH256 pKa = 7.42DD257 pKa = 4.38PSDD260 pKa = 4.14DD261 pKa = 4.32DD262 pKa = 5.47IYY264 pKa = 10.96IKK266 pKa = 10.94SKK268 pKa = 11.04GSFLSNLSGKK278 pKa = 7.76TKK280 pKa = 9.92IIKK283 pKa = 9.74RR284 pKa = 11.84RR285 pKa = 11.84TMRR288 pKa = 11.84AKK290 pKa = 10.51PIDD293 pKa = 3.74ITPADD298 pKa = 3.76HH299 pKa = 6.66NSNVRR304 pKa = 11.84SISSDD309 pKa = 2.85EE310 pKa = 4.01HH311 pKa = 7.81LGEE314 pKa = 4.18SEE316 pKa = 4.55RR317 pKa = 11.84RR318 pKa = 11.84SCSKK322 pKa = 10.41ISASDD327 pKa = 3.98CYY329 pKa = 9.89QTLNHH334 pKa = 5.89QAVLKK339 pKa = 10.25RR340 pKa = 11.84AMSSRR345 pKa = 11.84YY346 pKa = 9.13GG347 pKa = 3.31
MM1 pKa = 7.58CLSCLKK7 pKa = 10.28YY8 pKa = 10.72VWNLCLTMCDD18 pKa = 3.07LRR20 pKa = 11.84KK21 pKa = 9.62LVRR24 pKa = 11.84VTMINVNDD32 pKa = 3.49VRR34 pKa = 11.84KK35 pKa = 9.98QIIKK39 pKa = 10.51SGSLTSAVGTGYY51 pKa = 11.1VYY53 pKa = 10.84DD54 pKa = 4.05GSYY57 pKa = 11.05NKK59 pKa = 10.03YY60 pKa = 9.56ARR62 pKa = 11.84KK63 pKa = 9.7RR64 pKa = 11.84EE65 pKa = 3.97LNLRR69 pKa = 11.84VSSSGDD75 pKa = 3.14KK76 pKa = 10.56NIMMRR81 pKa = 11.84HH82 pKa = 4.69VPIFDD87 pKa = 5.11DD88 pKa = 3.67EE89 pKa = 4.4DD90 pKa = 3.6LRR92 pKa = 11.84ALRR95 pKa = 11.84SEE97 pKa = 4.29ANSNKK102 pKa = 9.82YY103 pKa = 9.21IHH105 pKa = 6.85IGCLTISIEE114 pKa = 4.05PLMHH118 pKa = 6.58KK119 pKa = 10.08RR120 pKa = 11.84YY121 pKa = 9.07MDD123 pKa = 4.17LYY125 pKa = 8.05GDD127 pKa = 3.9KK128 pKa = 10.17MRR130 pKa = 11.84GICAVIDD137 pKa = 3.52TTFADD142 pKa = 3.95PAEE145 pKa = 4.84SIISAHH151 pKa = 6.8RR152 pKa = 11.84YY153 pKa = 7.26EE154 pKa = 4.35LSKK157 pKa = 11.12GRR159 pKa = 11.84ADD161 pKa = 3.67FVSMPNHH168 pKa = 6.43CLSLLDD174 pKa = 4.42PNLKK178 pKa = 9.67QRR180 pKa = 11.84LSVMISLDD188 pKa = 3.9GINVKK193 pKa = 10.21KK194 pKa = 10.34GNEE197 pKa = 3.97MFNVCIGYY205 pKa = 9.47IVTGVNTLNPTKK217 pKa = 10.15AVSMSNIPITGTSEE231 pKa = 4.26CEE233 pKa = 3.9PGDD236 pKa = 3.69LSEE239 pKa = 6.36DD240 pKa = 3.29MLEE243 pKa = 4.66GIKK246 pKa = 10.69GSNGNLLISHH256 pKa = 7.42DD257 pKa = 4.38PSDD260 pKa = 4.14DD261 pKa = 4.32DD262 pKa = 5.47IYY264 pKa = 10.96IKK266 pKa = 10.94SKK268 pKa = 11.04GSFLSNLSGKK278 pKa = 7.76TKK280 pKa = 9.92IIKK283 pKa = 9.74RR284 pKa = 11.84RR285 pKa = 11.84TMRR288 pKa = 11.84AKK290 pKa = 10.51PIDD293 pKa = 3.74ITPADD298 pKa = 3.76HH299 pKa = 6.66NSNVRR304 pKa = 11.84SISSDD309 pKa = 2.85EE310 pKa = 4.01HH311 pKa = 7.81LGEE314 pKa = 4.18SEE316 pKa = 4.55RR317 pKa = 11.84RR318 pKa = 11.84SCSKK322 pKa = 10.41ISASDD327 pKa = 3.98CYY329 pKa = 9.89QTLNHH334 pKa = 5.89QAVLKK339 pKa = 10.25RR340 pKa = 11.84AMSSRR345 pKa = 11.84YY346 pKa = 9.13GG347 pKa = 3.31
Molecular weight: 38.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3919 |
168 |
2071 |
653.2 |
73.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.945 ± 0.953 | 1.863 ± 0.421 |
5.639 ± 0.466 | 5.588 ± 0.443 |
3.292 ± 0.431 | 5.537 ± 0.262 |
2.297 ± 0.152 | 6.405 ± 0.343 |
6.073 ± 0.543 | 10.564 ± 0.801 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.343 ± 0.196 | 4.925 ± 0.284 |
4.772 ± 0.274 | 2.679 ± 0.386 |
5.41 ± 0.417 | 8.497 ± 0.824 |
5.716 ± 0.444 | 6.634 ± 0.451 |
1.531 ± 0.286 | 3.292 ± 0.217 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
