
Alsobacter soli
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Alsobacteraceae; Alsobacter
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
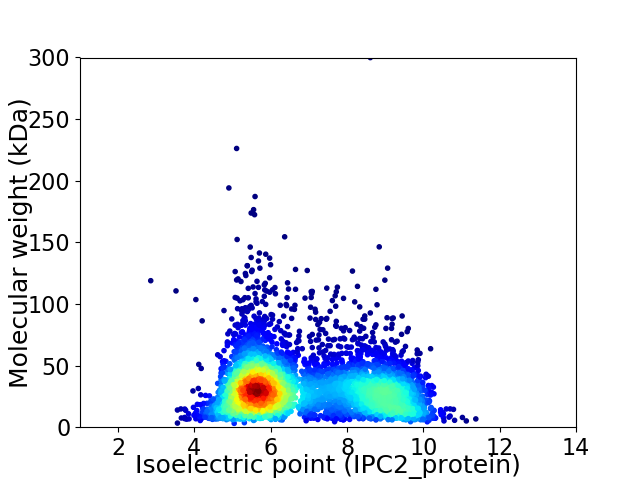
Virtual 2D-PAGE plot for 4752 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T1HX16|A0A2T1HX16_9RHIZ Lytic transglycosylase OS=Alsobacter soli OX=2109933 GN=SLNSH_04955 PE=4 SV=1
MM1 pKa = 7.95GDD3 pKa = 3.71TLSGGAGDD11 pKa = 4.13DD12 pKa = 4.46LIYY15 pKa = 10.94GSGGADD21 pKa = 3.31SLDD24 pKa = 3.58GGAGNDD30 pKa = 3.6TLVAGSGADD39 pKa = 3.54TLVGGAGVDD48 pKa = 3.48TADD51 pKa = 3.48YY52 pKa = 11.02SAFPTLISNGDD63 pKa = 3.59FSSAATGWTLGSNTAVTSGAAQATNATRR91 pKa = 11.84MLTQDD96 pKa = 2.81VGFVVGQTYY105 pKa = 7.93TVTFDD110 pKa = 3.39YY111 pKa = 11.19TMTAGARR118 pKa = 11.84LRR120 pKa = 11.84ISNSDD125 pKa = 3.32TNGANVVYY133 pKa = 9.72TSTVLDD139 pKa = 3.53SAGTVTATFVATGSKK154 pKa = 10.33LGIEE158 pKa = 4.39ADD160 pKa = 3.53SGSFTGTLDD169 pKa = 4.1NIRR172 pKa = 11.84MLQVVDD178 pKa = 4.29LATGKK183 pKa = 11.07SNDD186 pKa = 3.25GDD188 pKa = 3.67VLQGVEE194 pKa = 4.19NVIGGATGNKK204 pKa = 9.43IYY206 pKa = 10.86GDD208 pKa = 3.6TGANRR213 pKa = 11.84LDD215 pKa = 3.75GGAGNDD221 pKa = 3.65TLAGGSGADD230 pKa = 3.73TLAGGAGNDD239 pKa = 3.36RR240 pKa = 11.84LNGGEE245 pKa = 4.1AWLADD250 pKa = 4.06KK251 pKa = 10.92EE252 pKa = 4.36PASLTNAYY260 pKa = 10.09LFQNKK265 pKa = 7.57ATSFYY270 pKa = 10.59NGEE273 pKa = 4.17SLYY276 pKa = 10.56IPAQYY281 pKa = 8.73THH283 pKa = 7.05NIFEE287 pKa = 4.93HH288 pKa = 6.48PNSGAGEE295 pKa = 4.2TVAVYY300 pKa = 10.86NLGGATTFSSDD311 pKa = 4.52FVTNWSTSDD320 pKa = 3.13GATFRR325 pKa = 11.84VYY327 pKa = 11.34ADD329 pKa = 3.27GALIFEE335 pKa = 4.87KK336 pKa = 10.68VDD338 pKa = 3.48APSSQVFSTGLLNVAGASQLKK359 pKa = 10.18LVTTAGTSGNNGADD373 pKa = 2.93HH374 pKa = 7.11AMWLNPRR381 pKa = 11.84LSWGIADD388 pKa = 4.22GADD391 pKa = 4.03SISGGAGDD399 pKa = 4.3DD400 pKa = 3.97TIEE403 pKa = 4.48GGFGADD409 pKa = 3.51TLDD412 pKa = 3.89GGSGVDD418 pKa = 3.42TLDD421 pKa = 3.41YY422 pKa = 9.41TASTGAVTVNLATGAASGGDD442 pKa = 3.5AQGDD446 pKa = 3.76IISGFEE452 pKa = 4.14NVWGSAQADD461 pKa = 4.14SLVGDD466 pKa = 4.3AGANIFIGGLGADD479 pKa = 4.37TIVGGAGVDD488 pKa = 3.5TVDD491 pKa = 4.0YY492 pKa = 8.65STSGMNPAGGVGFFVDD508 pKa = 4.07LLAGKK513 pKa = 8.79GHH515 pKa = 6.63NGAAEE520 pKa = 4.23GDD522 pKa = 3.95VLSGIEE528 pKa = 3.99NLVGSVWNDD537 pKa = 3.06KK538 pKa = 11.08LYY540 pKa = 11.44GDD542 pKa = 4.77AGNNRR547 pKa = 11.84IDD549 pKa = 3.6GGGWGNDD556 pKa = 3.66LLSGGAGNDD565 pKa = 3.66TLIGGAGNEE574 pKa = 3.99RR575 pKa = 11.84LVGGLTWLADD585 pKa = 3.58IAPTSQTNILQYY597 pKa = 9.41STPGPGGLIPFQYY610 pKa = 9.48QHH612 pKa = 6.91NIFEE616 pKa = 5.1HH617 pKa = 6.3PNSTGEE623 pKa = 3.99AVLTYY628 pKa = 10.17DD629 pKa = 3.9IGSATTFSASFIKK642 pKa = 10.6NVSAGGDD649 pKa = 3.47GVTWRR654 pKa = 11.84VYY656 pKa = 11.2GDD658 pKa = 3.39STLLFEE664 pKa = 5.27MADD667 pKa = 3.46APLGQMFSTGVINVAGYY684 pKa = 8.91QQLKK688 pKa = 10.44LVTTSGSAGNNGFDD702 pKa = 3.24SAYY705 pKa = 9.02WLNPVLSSGAVGSDD719 pKa = 2.8NDD721 pKa = 4.36SLVGGAGNDD730 pKa = 3.72TLEE733 pKa = 4.68GGAGADD739 pKa = 3.86TMDD742 pKa = 4.38GGAGTDD748 pKa = 3.49TLDD751 pKa = 3.59YY752 pKa = 9.81AASSAAVTVDD762 pKa = 3.72LSANTASGGDD772 pKa = 3.48AQGDD776 pKa = 4.02VISGFEE782 pKa = 4.09YY783 pKa = 10.83VLGSAFADD791 pKa = 5.12RR792 pKa = 11.84ITGDD796 pKa = 3.92ANANWFYY803 pKa = 11.18GGSGDD808 pKa = 4.39DD809 pKa = 3.87TLIGAAGADD818 pKa = 3.58TMVGGWGADD827 pKa = 3.36SMDD830 pKa = 3.99GGSGVDD836 pKa = 3.27TLDD839 pKa = 3.44YY840 pKa = 9.82TGSSTGVTVNLATGVGLGGDD860 pKa = 3.69AQGDD864 pKa = 3.91TLIGFEE870 pKa = 4.49NVWGTSSADD879 pKa = 3.63SLVGDD884 pKa = 4.19ANDD887 pKa = 3.92NIFRR891 pKa = 11.84PYY893 pKa = 10.52LGADD897 pKa = 3.94TIVGGAGQDD906 pKa = 3.59TVDD909 pKa = 3.17YY910 pKa = 8.78WYY912 pKa = 10.88SGTGVSIDD920 pKa = 3.93LTLSTAQSGGDD931 pKa = 3.21AAGDD935 pKa = 3.46VFSGIEE941 pKa = 3.94NVIGSTTAANWIKK954 pKa = 11.15GDD956 pKa = 3.77ANANALTGWNGNDD969 pKa = 3.36TLIGGGGADD978 pKa = 3.9TLSGGAGNDD987 pKa = 3.07AFTLDD992 pKa = 3.94GSSLTIAAKK1001 pKa = 10.49VDD1003 pKa = 3.3GGTGADD1009 pKa = 3.55TLTIAANSGSSFTAANLAAAVTNMEE1034 pKa = 4.47VIDD1037 pKa = 3.87FRR1039 pKa = 11.84QAGVNANLSLSGAQVAAMTDD1059 pKa = 3.31ANKK1062 pKa = 10.76DD1063 pKa = 3.42LTINTSLGGGDD1074 pKa = 4.15TIAIADD1080 pKa = 3.67AASRR1084 pKa = 11.84YY1085 pKa = 7.39TMSVSGNTTNYY1096 pKa = 10.66DD1097 pKa = 3.02IYY1099 pKa = 11.35ADD1101 pKa = 4.16DD1102 pKa = 3.7AHH1104 pKa = 5.89TTLLAHH1110 pKa = 6.29LHH1112 pKa = 5.27VVAAA1116 pKa = 4.72
MM1 pKa = 7.95GDD3 pKa = 3.71TLSGGAGDD11 pKa = 4.13DD12 pKa = 4.46LIYY15 pKa = 10.94GSGGADD21 pKa = 3.31SLDD24 pKa = 3.58GGAGNDD30 pKa = 3.6TLVAGSGADD39 pKa = 3.54TLVGGAGVDD48 pKa = 3.48TADD51 pKa = 3.48YY52 pKa = 11.02SAFPTLISNGDD63 pKa = 3.59FSSAATGWTLGSNTAVTSGAAQATNATRR91 pKa = 11.84MLTQDD96 pKa = 2.81VGFVVGQTYY105 pKa = 7.93TVTFDD110 pKa = 3.39YY111 pKa = 11.19TMTAGARR118 pKa = 11.84LRR120 pKa = 11.84ISNSDD125 pKa = 3.32TNGANVVYY133 pKa = 9.72TSTVLDD139 pKa = 3.53SAGTVTATFVATGSKK154 pKa = 10.33LGIEE158 pKa = 4.39ADD160 pKa = 3.53SGSFTGTLDD169 pKa = 4.1NIRR172 pKa = 11.84MLQVVDD178 pKa = 4.29LATGKK183 pKa = 11.07SNDD186 pKa = 3.25GDD188 pKa = 3.67VLQGVEE194 pKa = 4.19NVIGGATGNKK204 pKa = 9.43IYY206 pKa = 10.86GDD208 pKa = 3.6TGANRR213 pKa = 11.84LDD215 pKa = 3.75GGAGNDD221 pKa = 3.65TLAGGSGADD230 pKa = 3.73TLAGGAGNDD239 pKa = 3.36RR240 pKa = 11.84LNGGEE245 pKa = 4.1AWLADD250 pKa = 4.06KK251 pKa = 10.92EE252 pKa = 4.36PASLTNAYY260 pKa = 10.09LFQNKK265 pKa = 7.57ATSFYY270 pKa = 10.59NGEE273 pKa = 4.17SLYY276 pKa = 10.56IPAQYY281 pKa = 8.73THH283 pKa = 7.05NIFEE287 pKa = 4.93HH288 pKa = 6.48PNSGAGEE295 pKa = 4.2TVAVYY300 pKa = 10.86NLGGATTFSSDD311 pKa = 4.52FVTNWSTSDD320 pKa = 3.13GATFRR325 pKa = 11.84VYY327 pKa = 11.34ADD329 pKa = 3.27GALIFEE335 pKa = 4.87KK336 pKa = 10.68VDD338 pKa = 3.48APSSQVFSTGLLNVAGASQLKK359 pKa = 10.18LVTTAGTSGNNGADD373 pKa = 2.93HH374 pKa = 7.11AMWLNPRR381 pKa = 11.84LSWGIADD388 pKa = 4.22GADD391 pKa = 4.03SISGGAGDD399 pKa = 4.3DD400 pKa = 3.97TIEE403 pKa = 4.48GGFGADD409 pKa = 3.51TLDD412 pKa = 3.89GGSGVDD418 pKa = 3.42TLDD421 pKa = 3.41YY422 pKa = 9.41TASTGAVTVNLATGAASGGDD442 pKa = 3.5AQGDD446 pKa = 3.76IISGFEE452 pKa = 4.14NVWGSAQADD461 pKa = 4.14SLVGDD466 pKa = 4.3AGANIFIGGLGADD479 pKa = 4.37TIVGGAGVDD488 pKa = 3.5TVDD491 pKa = 4.0YY492 pKa = 8.65STSGMNPAGGVGFFVDD508 pKa = 4.07LLAGKK513 pKa = 8.79GHH515 pKa = 6.63NGAAEE520 pKa = 4.23GDD522 pKa = 3.95VLSGIEE528 pKa = 3.99NLVGSVWNDD537 pKa = 3.06KK538 pKa = 11.08LYY540 pKa = 11.44GDD542 pKa = 4.77AGNNRR547 pKa = 11.84IDD549 pKa = 3.6GGGWGNDD556 pKa = 3.66LLSGGAGNDD565 pKa = 3.66TLIGGAGNEE574 pKa = 3.99RR575 pKa = 11.84LVGGLTWLADD585 pKa = 3.58IAPTSQTNILQYY597 pKa = 9.41STPGPGGLIPFQYY610 pKa = 9.48QHH612 pKa = 6.91NIFEE616 pKa = 5.1HH617 pKa = 6.3PNSTGEE623 pKa = 3.99AVLTYY628 pKa = 10.17DD629 pKa = 3.9IGSATTFSASFIKK642 pKa = 10.6NVSAGGDD649 pKa = 3.47GVTWRR654 pKa = 11.84VYY656 pKa = 11.2GDD658 pKa = 3.39STLLFEE664 pKa = 5.27MADD667 pKa = 3.46APLGQMFSTGVINVAGYY684 pKa = 8.91QQLKK688 pKa = 10.44LVTTSGSAGNNGFDD702 pKa = 3.24SAYY705 pKa = 9.02WLNPVLSSGAVGSDD719 pKa = 2.8NDD721 pKa = 4.36SLVGGAGNDD730 pKa = 3.72TLEE733 pKa = 4.68GGAGADD739 pKa = 3.86TMDD742 pKa = 4.38GGAGTDD748 pKa = 3.49TLDD751 pKa = 3.59YY752 pKa = 9.81AASSAAVTVDD762 pKa = 3.72LSANTASGGDD772 pKa = 3.48AQGDD776 pKa = 4.02VISGFEE782 pKa = 4.09YY783 pKa = 10.83VLGSAFADD791 pKa = 5.12RR792 pKa = 11.84ITGDD796 pKa = 3.92ANANWFYY803 pKa = 11.18GGSGDD808 pKa = 4.39DD809 pKa = 3.87TLIGAAGADD818 pKa = 3.58TMVGGWGADD827 pKa = 3.36SMDD830 pKa = 3.99GGSGVDD836 pKa = 3.27TLDD839 pKa = 3.44YY840 pKa = 9.82TGSSTGVTVNLATGVGLGGDD860 pKa = 3.69AQGDD864 pKa = 3.91TLIGFEE870 pKa = 4.49NVWGTSSADD879 pKa = 3.63SLVGDD884 pKa = 4.19ANDD887 pKa = 3.92NIFRR891 pKa = 11.84PYY893 pKa = 10.52LGADD897 pKa = 3.94TIVGGAGQDD906 pKa = 3.59TVDD909 pKa = 3.17YY910 pKa = 8.78WYY912 pKa = 10.88SGTGVSIDD920 pKa = 3.93LTLSTAQSGGDD931 pKa = 3.21AAGDD935 pKa = 3.46VFSGIEE941 pKa = 3.94NVIGSTTAANWIKK954 pKa = 11.15GDD956 pKa = 3.77ANANALTGWNGNDD969 pKa = 3.36TLIGGGGADD978 pKa = 3.9TLSGGAGNDD987 pKa = 3.07AFTLDD992 pKa = 3.94GSSLTIAAKK1001 pKa = 10.49VDD1003 pKa = 3.3GGTGADD1009 pKa = 3.55TLTIAANSGSSFTAANLAAAVTNMEE1034 pKa = 4.47VIDD1037 pKa = 3.87FRR1039 pKa = 11.84QAGVNANLSLSGAQVAAMTDD1059 pKa = 3.31ANKK1062 pKa = 10.76DD1063 pKa = 3.42LTINTSLGGGDD1074 pKa = 4.15TIAIADD1080 pKa = 3.67AASRR1084 pKa = 11.84YY1085 pKa = 7.39TMSVSGNTTNYY1096 pKa = 10.66DD1097 pKa = 3.02IYY1099 pKa = 11.35ADD1101 pKa = 4.16DD1102 pKa = 3.7AHH1104 pKa = 5.89TTLLAHH1110 pKa = 6.29LHH1112 pKa = 5.27VVAAA1116 pKa = 4.72
Molecular weight: 110.57 kDa
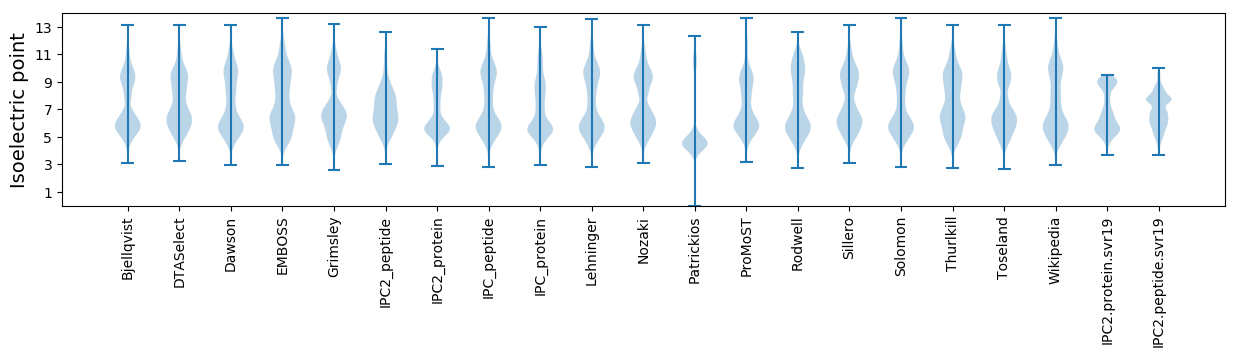
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T1HRP6|A0A2T1HRP6_9RHIZ 30S ribosomal protein S4 OS=Alsobacter soli OX=2109933 GN=rpsD PE=3 SV=1
AA1 pKa = 7.47GRR3 pKa = 11.84GGGGGAARR11 pKa = 11.84SGWTGLATGSGRR23 pKa = 11.84AGRR26 pKa = 11.84AGSGRR31 pKa = 11.84ASARR35 pKa = 11.84STGSGRR41 pKa = 11.84IAGSGRR47 pKa = 11.84RR48 pKa = 11.84TGSARR53 pKa = 11.84RR54 pKa = 11.84SGSGRR59 pKa = 11.84ITSGRR64 pKa = 11.84RR65 pKa = 11.84GGGGGGLRR73 pKa = 4.03
AA1 pKa = 7.47GRR3 pKa = 11.84GGGGGAARR11 pKa = 11.84SGWTGLATGSGRR23 pKa = 11.84AGRR26 pKa = 11.84AGSGRR31 pKa = 11.84ASARR35 pKa = 11.84STGSGRR41 pKa = 11.84IAGSGRR47 pKa = 11.84RR48 pKa = 11.84TGSARR53 pKa = 11.84RR54 pKa = 11.84SGSGRR59 pKa = 11.84ITSGRR64 pKa = 11.84RR65 pKa = 11.84GGGGGGLRR73 pKa = 4.03
Molecular weight: 6.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1490446 |
30 |
2818 |
313.6 |
33.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.787 ± 0.051 | 0.839 ± 0.01 |
5.387 ± 0.031 | 5.6 ± 0.034 |
3.559 ± 0.022 | 8.931 ± 0.038 |
1.884 ± 0.018 | 4.421 ± 0.028 |
3.042 ± 0.031 | 10.246 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.353 ± 0.016 | 2.221 ± 0.02 |
5.635 ± 0.027 | 3.067 ± 0.022 |
7.669 ± 0.04 | 5.154 ± 0.025 |
4.898 ± 0.023 | 7.897 ± 0.036 |
1.343 ± 0.015 | 2.065 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
