
Candidatus Marinimicrobia bacterium MT.SAG.3
Taxonomy: cellular organisms; Bacteria; FCB group; Candidatus Marinimicrobia
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
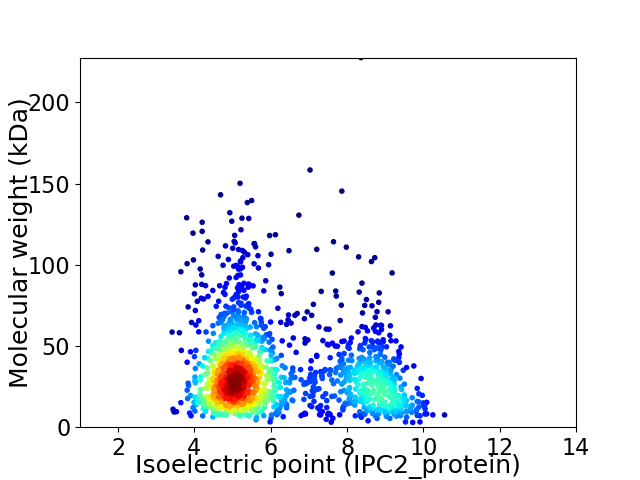
Virtual 2D-PAGE plot for 1570 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y8IED9|A0A4Y8IED9_9BACT Uncharacterized protein OS=Candidatus Marinimicrobia bacterium MT.SAG.3 OX=2552982 GN=E3V55_02365 PE=4 SV=1
MM1 pKa = 7.8LKK3 pKa = 9.07RR4 pKa = 11.84TLLLTLTICAVASLAIPQPAYY25 pKa = 10.58AQISEE30 pKa = 4.32FKK32 pKa = 9.49ITAADD37 pKa = 3.95GAAGDD42 pKa = 4.05LFGNSVSISGDD53 pKa = 3.46YY54 pKa = 10.87AVVGARR60 pKa = 11.84RR61 pKa = 11.84DD62 pKa = 3.64DD63 pKa = 4.25DD64 pKa = 4.33NGISSGSAYY73 pKa = 10.18VYY75 pKa = 10.84KK76 pKa = 10.02RR77 pKa = 11.84TGTSWAQEE85 pKa = 4.02AKK87 pKa = 10.77LLASDD92 pKa = 5.04GAADD96 pKa = 5.72DD97 pKa = 4.19LFGWSVSISGDD108 pKa = 3.57YY109 pKa = 10.87AVVGARR115 pKa = 11.84RR116 pKa = 11.84DD117 pKa = 3.64DD118 pKa = 4.25DD119 pKa = 4.33NGISSGSAYY128 pKa = 10.18VYY130 pKa = 10.84KK131 pKa = 10.02RR132 pKa = 11.84TGTSWAQEE140 pKa = 4.02AKK142 pKa = 10.77LLASDD147 pKa = 5.04GAADD151 pKa = 5.72DD152 pKa = 4.19LFGWSVSISGDD163 pKa = 3.63YY164 pKa = 10.9AVVGAFGDD172 pKa = 4.14DD173 pKa = 4.65DD174 pKa = 4.58NGTDD178 pKa = 3.27AGSAYY183 pKa = 10.15VFKK186 pKa = 10.55RR187 pKa = 11.84TGTSWAQEE195 pKa = 4.02AKK197 pKa = 10.73LLASDD202 pKa = 4.69SGVGDD207 pKa = 3.26QFGNSVSISGDD218 pKa = 3.59YY219 pKa = 10.88AVVGAWLDD227 pKa = 3.86DD228 pKa = 4.11DD229 pKa = 4.95SGSASGSAYY238 pKa = 10.72LFTRR242 pKa = 11.84SGTSWAQKK250 pKa = 10.7AKK252 pKa = 10.79LLASDD257 pKa = 4.58GATEE261 pKa = 4.25GFFGKK266 pKa = 10.02SVSISGDD273 pKa = 3.44YY274 pKa = 10.93AIVGAPAEE282 pKa = 4.5NPGSAYY288 pKa = 10.43LYY290 pKa = 10.59YY291 pKa = 11.03GFTFPQEE298 pKa = 3.76VAMLSFVVEE307 pKa = 3.92PVYY310 pKa = 10.7AVPDD314 pKa = 3.76SDD316 pKa = 4.02SLKK319 pKa = 10.85VNALMASSSGLTLMAAFEE337 pKa = 4.66SVDD340 pKa = 3.58SVLIDD345 pKa = 3.57SVYY348 pKa = 10.99LFDD351 pKa = 6.89DD352 pKa = 4.38DD353 pKa = 4.49AHH355 pKa = 7.01NDD357 pKa = 4.25GIAGDD362 pKa = 4.16SLFGNLWLVPAEE374 pKa = 3.94EE375 pKa = 3.49RR376 pKa = 11.84HH377 pKa = 5.55YY378 pKa = 10.96LVNIHH383 pKa = 5.78VSKK386 pKa = 11.06DD387 pKa = 3.03DD388 pKa = 3.32TLIYY392 pKa = 10.18IRR394 pKa = 11.84EE395 pKa = 3.85NAARR399 pKa = 11.84FTTIGPVVLDD409 pKa = 4.09SYY411 pKa = 10.71TLFDD415 pKa = 4.75TIPNPGEE422 pKa = 3.85RR423 pKa = 11.84LFLEE427 pKa = 3.91LTLRR431 pKa = 11.84NNGSTANATAITADD445 pKa = 3.05ISTSDD450 pKa = 3.38TCVTDD455 pKa = 4.41IGALGQNTYY464 pKa = 11.43GDD466 pKa = 3.92IGPGATATSAGAYY479 pKa = 9.1IVDD482 pKa = 3.93VNEE485 pKa = 4.13NCAGDD490 pKa = 3.82QDD492 pKa = 3.94ILFDD496 pKa = 3.97ISIASEE502 pKa = 4.69GFDD505 pKa = 3.22FWGDD509 pKa = 3.08SFTIHH514 pKa = 6.48VFPTDD519 pKa = 3.11TTIPQTRR526 pKa = 11.84WHH528 pKa = 6.68EE529 pKa = 3.96KK530 pKa = 10.64SIDD533 pKa = 3.82SVDD536 pKa = 3.5GNFSQGVATDD546 pKa = 3.73GVDD549 pKa = 2.88WYY551 pKa = 10.86FSARR555 pKa = 11.84QTLFKK560 pKa = 10.6TNYY563 pKa = 9.85DD564 pKa = 3.29YY565 pKa = 11.88DD566 pKa = 3.91FLVVNEE572 pKa = 4.06NPIPQFLQDD581 pKa = 3.25QSYY584 pKa = 11.34DD585 pKa = 3.71HH586 pKa = 7.23IGDD589 pKa = 3.23IAYY592 pKa = 10.41FEE594 pKa = 4.6GKK596 pKa = 10.02LYY598 pKa = 11.02APIEE602 pKa = 4.38DD603 pKa = 3.51ASYY606 pKa = 10.75VKK608 pKa = 10.25PIICLYY614 pKa = 10.72DD615 pKa = 3.59AVSLDD620 pKa = 3.55FTGEE624 pKa = 4.16FAQVPQSHH632 pKa = 6.52IPWVAVDD639 pKa = 3.44PRR641 pKa = 11.84TGYY644 pKa = 10.36FYY646 pKa = 10.9SSEE649 pKa = 4.07FNGVNNLFVYY659 pKa = 10.44DD660 pKa = 4.17PNQNFALIDD669 pKa = 3.87EE670 pKa = 4.68VQLDD674 pKa = 3.98TTLSNVQGGAFLGDD688 pKa = 3.8FLYY691 pKa = 10.96LAA693 pKa = 5.06
MM1 pKa = 7.8LKK3 pKa = 9.07RR4 pKa = 11.84TLLLTLTICAVASLAIPQPAYY25 pKa = 10.58AQISEE30 pKa = 4.32FKK32 pKa = 9.49ITAADD37 pKa = 3.95GAAGDD42 pKa = 4.05LFGNSVSISGDD53 pKa = 3.46YY54 pKa = 10.87AVVGARR60 pKa = 11.84RR61 pKa = 11.84DD62 pKa = 3.64DD63 pKa = 4.25DD64 pKa = 4.33NGISSGSAYY73 pKa = 10.18VYY75 pKa = 10.84KK76 pKa = 10.02RR77 pKa = 11.84TGTSWAQEE85 pKa = 4.02AKK87 pKa = 10.77LLASDD92 pKa = 5.04GAADD96 pKa = 5.72DD97 pKa = 4.19LFGWSVSISGDD108 pKa = 3.57YY109 pKa = 10.87AVVGARR115 pKa = 11.84RR116 pKa = 11.84DD117 pKa = 3.64DD118 pKa = 4.25DD119 pKa = 4.33NGISSGSAYY128 pKa = 10.18VYY130 pKa = 10.84KK131 pKa = 10.02RR132 pKa = 11.84TGTSWAQEE140 pKa = 4.02AKK142 pKa = 10.77LLASDD147 pKa = 5.04GAADD151 pKa = 5.72DD152 pKa = 4.19LFGWSVSISGDD163 pKa = 3.63YY164 pKa = 10.9AVVGAFGDD172 pKa = 4.14DD173 pKa = 4.65DD174 pKa = 4.58NGTDD178 pKa = 3.27AGSAYY183 pKa = 10.15VFKK186 pKa = 10.55RR187 pKa = 11.84TGTSWAQEE195 pKa = 4.02AKK197 pKa = 10.73LLASDD202 pKa = 4.69SGVGDD207 pKa = 3.26QFGNSVSISGDD218 pKa = 3.59YY219 pKa = 10.88AVVGAWLDD227 pKa = 3.86DD228 pKa = 4.11DD229 pKa = 4.95SGSASGSAYY238 pKa = 10.72LFTRR242 pKa = 11.84SGTSWAQKK250 pKa = 10.7AKK252 pKa = 10.79LLASDD257 pKa = 4.58GATEE261 pKa = 4.25GFFGKK266 pKa = 10.02SVSISGDD273 pKa = 3.44YY274 pKa = 10.93AIVGAPAEE282 pKa = 4.5NPGSAYY288 pKa = 10.43LYY290 pKa = 10.59YY291 pKa = 11.03GFTFPQEE298 pKa = 3.76VAMLSFVVEE307 pKa = 3.92PVYY310 pKa = 10.7AVPDD314 pKa = 3.76SDD316 pKa = 4.02SLKK319 pKa = 10.85VNALMASSSGLTLMAAFEE337 pKa = 4.66SVDD340 pKa = 3.58SVLIDD345 pKa = 3.57SVYY348 pKa = 10.99LFDD351 pKa = 6.89DD352 pKa = 4.38DD353 pKa = 4.49AHH355 pKa = 7.01NDD357 pKa = 4.25GIAGDD362 pKa = 4.16SLFGNLWLVPAEE374 pKa = 3.94EE375 pKa = 3.49RR376 pKa = 11.84HH377 pKa = 5.55YY378 pKa = 10.96LVNIHH383 pKa = 5.78VSKK386 pKa = 11.06DD387 pKa = 3.03DD388 pKa = 3.32TLIYY392 pKa = 10.18IRR394 pKa = 11.84EE395 pKa = 3.85NAARR399 pKa = 11.84FTTIGPVVLDD409 pKa = 4.09SYY411 pKa = 10.71TLFDD415 pKa = 4.75TIPNPGEE422 pKa = 3.85RR423 pKa = 11.84LFLEE427 pKa = 3.91LTLRR431 pKa = 11.84NNGSTANATAITADD445 pKa = 3.05ISTSDD450 pKa = 3.38TCVTDD455 pKa = 4.41IGALGQNTYY464 pKa = 11.43GDD466 pKa = 3.92IGPGATATSAGAYY479 pKa = 9.1IVDD482 pKa = 3.93VNEE485 pKa = 4.13NCAGDD490 pKa = 3.82QDD492 pKa = 3.94ILFDD496 pKa = 3.97ISIASEE502 pKa = 4.69GFDD505 pKa = 3.22FWGDD509 pKa = 3.08SFTIHH514 pKa = 6.48VFPTDD519 pKa = 3.11TTIPQTRR526 pKa = 11.84WHH528 pKa = 6.68EE529 pKa = 3.96KK530 pKa = 10.64SIDD533 pKa = 3.82SVDD536 pKa = 3.5GNFSQGVATDD546 pKa = 3.73GVDD549 pKa = 2.88WYY551 pKa = 10.86FSARR555 pKa = 11.84QTLFKK560 pKa = 10.6TNYY563 pKa = 9.85DD564 pKa = 3.29YY565 pKa = 11.88DD566 pKa = 3.91FLVVNEE572 pKa = 4.06NPIPQFLQDD581 pKa = 3.25QSYY584 pKa = 11.34DD585 pKa = 3.71HH586 pKa = 7.23IGDD589 pKa = 3.23IAYY592 pKa = 10.41FEE594 pKa = 4.6GKK596 pKa = 10.02LYY598 pKa = 11.02APIEE602 pKa = 4.38DD603 pKa = 3.51ASYY606 pKa = 10.75VKK608 pKa = 10.25PIICLYY614 pKa = 10.72DD615 pKa = 3.59AVSLDD620 pKa = 3.55FTGEE624 pKa = 4.16FAQVPQSHH632 pKa = 6.52IPWVAVDD639 pKa = 3.44PRR641 pKa = 11.84TGYY644 pKa = 10.36FYY646 pKa = 10.9SSEE649 pKa = 4.07FNGVNNLFVYY659 pKa = 10.44DD660 pKa = 4.17PNQNFALIDD669 pKa = 3.87EE670 pKa = 4.68VQLDD674 pKa = 3.98TTLSNVQGGAFLGDD688 pKa = 3.8FLYY691 pKa = 10.96LAA693 pKa = 5.06
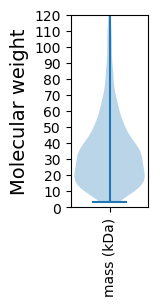
Molecular weight: 74.06 kDa
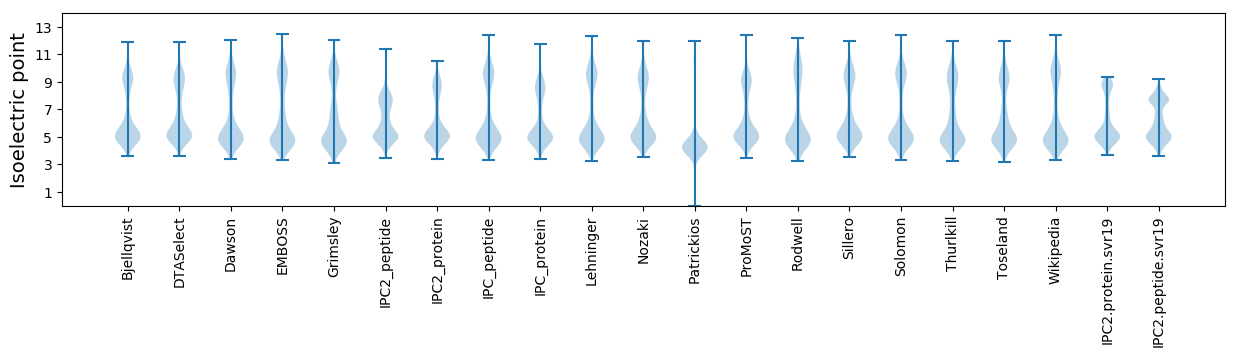
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y8I424|A0A4Y8I424_9BACT Uncharacterized protein OS=Candidatus Marinimicrobia bacterium MT.SAG.3 OX=2552982 GN=E3V55_05775 PE=4 SV=1
MM1 pKa = 7.6AKK3 pKa = 10.02KK4 pKa = 10.25SWIAKK9 pKa = 7.53QKK11 pKa = 8.76KK12 pKa = 4.13TQKK15 pKa = 10.16FAVRR19 pKa = 11.84EE20 pKa = 3.92YY21 pKa = 11.02NRR23 pKa = 11.84CQRR26 pKa = 11.84CGRR29 pKa = 11.84ARR31 pKa = 11.84SYY33 pKa = 10.81YY34 pKa = 10.18RR35 pKa = 11.84RR36 pKa = 11.84FGLCRR41 pKa = 11.84ICFRR45 pKa = 11.84EE46 pKa = 3.87LALKK50 pKa = 10.94GEE52 pKa = 4.37IPGITKK58 pKa = 10.6ASWW61 pKa = 2.73
MM1 pKa = 7.6AKK3 pKa = 10.02KK4 pKa = 10.25SWIAKK9 pKa = 7.53QKK11 pKa = 8.76KK12 pKa = 4.13TQKK15 pKa = 10.16FAVRR19 pKa = 11.84EE20 pKa = 3.92YY21 pKa = 11.02NRR23 pKa = 11.84CQRR26 pKa = 11.84CGRR29 pKa = 11.84ARR31 pKa = 11.84SYY33 pKa = 10.81YY34 pKa = 10.18RR35 pKa = 11.84RR36 pKa = 11.84FGLCRR41 pKa = 11.84ICFRR45 pKa = 11.84EE46 pKa = 3.87LALKK50 pKa = 10.94GEE52 pKa = 4.37IPGITKK58 pKa = 10.6ASWW61 pKa = 2.73
Molecular weight: 7.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
510553 |
25 |
2012 |
325.2 |
36.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.711 ± 0.056 | 0.61 ± 0.017 |
5.897 ± 0.052 | 7.316 ± 0.074 |
4.574 ± 0.05 | 7.368 ± 0.061 |
1.669 ± 0.022 | 8.134 ± 0.057 |
6.605 ± 0.072 | 9.759 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.497 ± 0.035 | 4.598 ± 0.044 |
3.639 ± 0.034 | 2.48 ± 0.026 |
4.587 ± 0.046 | 7.418 ± 0.054 |
5.173 ± 0.047 | 6.434 ± 0.045 |
1.065 ± 0.023 | 3.467 ± 0.043 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
